| Sequence ID | dm3.chr3L |
|---|---|
| Location | 832,489 – 832,596 |
| Length | 107 |
| Max. P | 0.679474 |

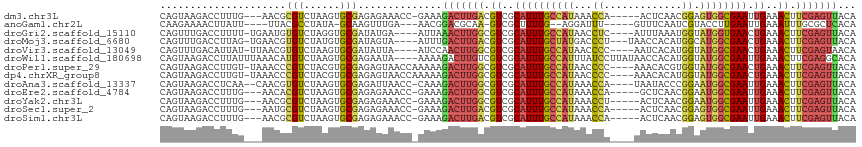
| Location | 832,489 – 832,596 |
|---|---|
| Length | 107 |
| Sequences | 13 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 75.99 |
| Shannon entropy | 0.51348 |
| G+C content | 0.43401 |
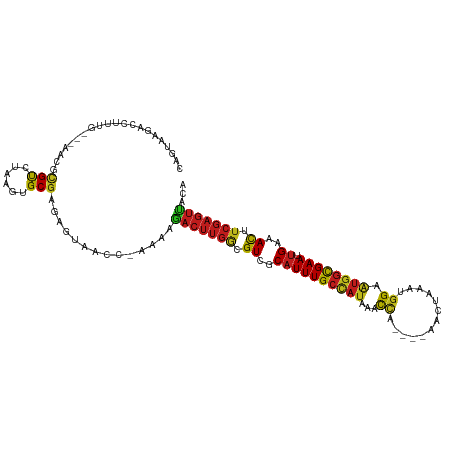
| Mean single sequence MFE | -25.04 |
| Consensus MFE | -15.64 |
| Energy contribution | -15.43 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.46 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.679474 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 832489 107 - 24543557 CAGUAAGACCUUUG---AACGCGUCUAAGUGCGAGAGAAACC-GAAAGACUUGACGUCGCAUUUGCCAUAAACCA-----ACUCAACGGAGUGGCGAAUUGAAACUUCGAGUUACA ..(((....((((.---..((((......))))))))....(-....)((((((.((..((((((((((...((.-----.......)).)))))))).))..)).))))))))). ( -25.00, z-score = -0.59, R) >anoGam1.chr2L 32405827 100 + 48795086 CAAGAAAACUUAUU----UUACGCCUAUA-GCAAGUUUGA---AACGGACGCAA-GUCGCUUUUG--AGGAUUU-----GUUUCAAUCGUACCUUGAAUUGAAAUUUGCGCUCACA ..............----...........-((((((((.(---(...(((....-)))...((..--(((...(-----(.......))..)))..)))).))))))))....... ( -19.50, z-score = -0.15, R) >droGri2.scaffold_15110 20029707 107 + 24565398 CAGUUUGACCUUUU-UGAAUGUGUCUAGGUGCGAUAUGA----AUUAAACUUGGCGUCGCAUUUGCCAUAACCUC----AUUUAAAUGGUAUGGUGAACUGAAACUUCGAGUUACA ((((((........-.(((((((.((((((..(((....----)))..))))))...)))))))((((((.((..----........))))))))))))))............... ( -24.70, z-score = -1.03, R) >droMoj3.scaffold_6680 6557277 108 - 24764193 CAGUUUGACCUUAG-UGAACGUGUCUAUGUGCGAUAGUA----AUUUGACUUGACGUCGCAUUUGCUAUGACCCU---UAACCACAUGGCAUGGCGAACUGAAACUUCGAGUUACA (((((((.((...(-(((.((((((....(((....)))----....)))...)))))))...(((((((.....---......))))))).)))))))))............... ( -28.20, z-score = -1.59, R) >droVir3.scaffold_13049 16449741 107 + 25233164 CAGUUUGACAUUAU-UUAACGUGUCUAAGUGCGAUAUUA----AUCCAACUUGGCGUCGCAUUUGCCAUAACCCC----AAUCACAUGGUAUGGCGAACUGAAACUUCGAGUAACA (((((((((((...-.....))))).((((((((..(((----(......))))..))))))))((((((...((----(......)))))))))))))))............... ( -29.30, z-score = -2.64, R) >droWil1.scaffold_180698 9069256 112 - 11422946 CAGUAAGACCUUAUUUAAACAUGUCUAAGUGCGAGAAUA----AAAAGACUUGUCGUCGCAUUUGCCAUUUAUCCUUAUAACCACAUGGUAUGGCGAAUUGAAACUUCGAGGCACA .....................(((((((((((((..(((----(......))))..))))(((((((((....((............)).)))))))))....))))..))))).. ( -23.00, z-score = -0.76, R) >droPer1.super_29 464414 111 + 1099123 CAGUAAGACCUUGU-UAAACCCGUCUACGUGCGAGAGUAACCAAAAAGACUUGGCGUCGCAUUUGCCAUAACCCC----AAACACGUGGUAUGGCGAACUGAAACUUCGAGUUACA ..(((.(((.....-.......)))))).(((....)))........(((((((.((..(((((((((((.((.(----......).))))))))))).))..)).)))))))... ( -27.70, z-score = -1.18, R) >dp4.chrXR_group8 4812631 111 + 9212921 CAGUAAGACCUUGU-UAAACCCGUCUACGUGCGAGAGUAACCAAAAAGACUUGGCGUCGCAUUUGCCAUAACCCC----AAACACAUGGUAUGGCGAACUGAAACUUCGAGUUACA ..(((.(((.....-.......)))))).(((....)))........(((((((.((..(((((((((((...((----(......)))))))))))).))..)).)))))))... ( -27.00, z-score = -1.35, R) >droAna3.scaffold_13337 11799252 109 - 23293914 CAGUAAGACCUCAA--CAACGUGUCUAAGUGCGAGAUUAACC-CAAAGACUUGGCGUCGCAUUUGCCAUAAACCA----UAAUACCCGGAAUGGCGAAUUGAAACUUCGAGUUACA ((.(.(((((....--....).)))).).))...........-....(((((((.((..((((((((((...((.----........)).)))))))).))..)).)))))))... ( -22.20, z-score = -0.51, R) >droEre2.scaffold_4784 848076 107 - 25762168 CAGUAAGACCUUUG---AACACGUCUAAGUGCGAGAGAAACC-GAAAGACUUGGCGUCGCAUUUGCCAUAAACCA-----GCUCAACGGAAUGGCGAAUUGAAACUUCGAGUUACA ..((((....((((---((..((.((((((.(....)....(-....)))))))))(((.(((((((((...((.-----.......)).))))))))))))...)))))))))). ( -24.30, z-score = -0.38, R) >droYak2.chr3L 828212 107 - 24197627 CAGUAAGACCUUUG---AACGCGUCUAAGUGCGAGAGAAACC-GAAAGACUUGGCGUCGCAUUUGCCAUAAACCU-----ACUCAACGGAAUGGCGAAUUGAAACUUCGAGUUACA ..((((....((((---((.(((.((((((.(....)....(-....)))))))...)))(((((((((...((.-----.......)).)))))))))......)))))))))). ( -25.00, z-score = -0.56, R) >droSec1.super_2 860318 107 - 7591821 CAGUAAGACCUUUG---AAUGCGUCUAAGUGCGAGAGAAACC-GAAAGACUUGACGUCGCAUUUGCCAUAAACCA-----ACUCAACGGAGUGGCGAAUUGAAACUUCGAGUUACA ..((((....((((---((..((((.((((.(....)....(-....)))))))))(((.(((((((((...((.-----.......)).))))))))))))...)))))))))). ( -24.60, z-score = -0.65, R) >droSim1.chr3L 487379 107 - 22553184 CAGUAAGACCUUUG---AACGCGUCUAAGUGCGAGAGAAACC-GAAAGACUUGACGUCGCAUUUGCCAUAAACCA-----ACUCAACGGAGUGGCGAAUUGAAACUUCGAGUUACA ..(((....((((.---..((((......))))))))....(-....)((((((.((..((((((((((...((.-----.......)).)))))))).))..)).))))))))). ( -25.00, z-score = -0.59, R) >consensus CAGUAAGACCUUUG___AACGCGUCUAAGUGCGAGAGUAACC_AAAAGACUUGGCGUCGCAUUUGCCAUAAACCA____AACUAAAUGGAAUGGCGAAUUGAAACUUCGAGUUACA .....................(((......)))..............(((((((.((..((((((((((...((.............)).)))))))).))..)).)))))))... (-15.64 = -15.43 + -0.21)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:54:29 2011