| Sequence ID | dm3.chr3L |
|---|---|
| Location | 831,929 – 832,036 |
| Length | 107 |
| Max. P | 0.998542 |

| Location | 831,929 – 832,019 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 64.58 |
| Shannon entropy | 0.72454 |
| G+C content | 0.43553 |
| Mean single sequence MFE | -23.95 |
| Consensus MFE | -12.15 |
| Energy contribution | -11.28 |
| Covariance contribution | -0.87 |
| Combinations/Pair | 1.69 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.39 |
| SVM RNA-class probability | 0.998542 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
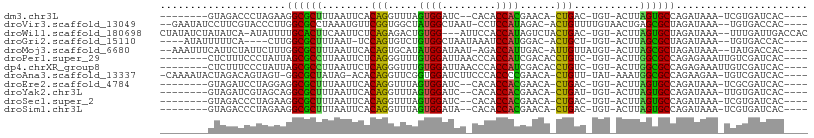
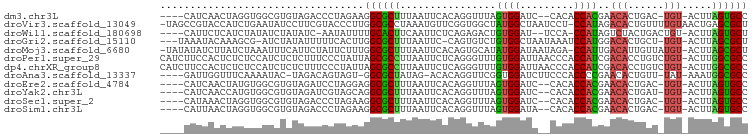
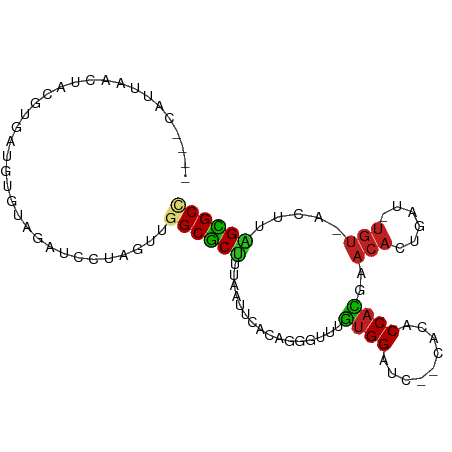
>dm3.chr3L 831929 90 - 24543557 --------GUAGACCCUAGAAGGCGCUUUAAUUCACAGGUUUAGUGGAUC--CACACCACGAACA-CUGAC-UGU-ACUUAGUGCCAGAUAAA-UCGUGAUCAC---- --------.............((((((........(((((((.((((...--....)))))))).-)))..-...-....)))))).(((...-.....)))..---- ( -19.93, z-score = -0.63, R) >droVir3.scaffold_13049 16449180 98 + 25233164 --GAAUAUCCUUCGUACCCUUGGCGCCUAAAUGUUCGGUGGCUAUGGCUAAU-CCUCCAUAGAC-ACUGUUUUGUAACUGAGCGCUAGAUAAA--UGUGACCAC---- --.................((((((((........(((((.((((((.....-...)))))).)-))))..........).))))))).....--.........---- ( -22.27, z-score = -0.75, R) >droWil1.scaffold_180698 9068687 100 - 11422946 CUAUAUCUAUAUCA-AUAUUUUGCACUUCAAUUCUCAGAGACUGUGG---AUUCCACCAUAGUCUACUGAC-UGU-ACUUAGUGCUAGAUAAA--UUUGAUUGACCAC .....((...((((-((((((.(((((...((..(((((((((((((---......))))))))).)))).-.))-....))))).)))))..--.))))).)).... ( -29.90, z-score = -4.86, R) >droGri2.scaffold_15110 20029140 90 + 24565398 ----AUAUUUUUCA----CUUGGCGCUUUAAU-UCCAGUGUCUGUGGCUAAUAAAUCCAUGGAC-ACUGCU-UGU-ACUUAGCGCUAGAUAAA--UGUGACCAC---- ----((((((.((.----..(((((((.((..-..((((((((((((.........))))))))-))))..-..)-)...))))))))).)))--)))......---- ( -28.90, z-score = -4.24, R) >droMoj3.scaffold_6680 6556722 97 - 24764193 --AAAUUUCAUUCUAUUCUUUGGCGCUUUAAUUCACAGUGCAUAUGGAUAAU-AGACCAUUGAC-AUUGUUAUGU-ACUUAGCGCUAGAUAAA--UAUGACCAC---- --.....((((.......(((((((((.((....((((((((.((((.....-...)))))).)-)))))....)-)...)))))))))....--.))))....---- ( -22.20, z-score = -2.53, R) >droPer1.super_29 463841 94 + 1099123 --------CUCUUUCCCUAUUAGCGCCUUAAUUCUCAGGGUUUGUGGAUUAACCCACCAUCGACACCUGUC-UGU-ACUUGGCGCCAGAGAAAUUGUCGAUCAC---- --------...((((.((....(((((........(((((((.((((.........)))).))).))))..-...-....))))).)).))))...........---- ( -22.83, z-score = -1.39, R) >dp4.chrXR_group8 4812058 94 + 9212921 --------CUCUUUCCCUAUUAGCGCCUUAAUUCUCAGGGUUUGUGGAUUAACCCACCAUCGACACCUGUC-UGU-ACUUGGCGCCAGAGAAAUUGUCGAUCAC---- --------...((((.((....(((((........(((((((.((((.........)))).))).))))..-...-....))))).)).))))...........---- ( -22.83, z-score = -1.39, R) >droAna3.scaffold_13337 11798697 97 - 23293914 -CAAAAUACUAGACAGUAGU-GGCGCUAUAG-ACACAGGUUCGGUGGAUCUUCCCACCCCGAACA-CUGUU-UAU-AAAUGGCGCCAGAAGAA-UGUCGAUCAC---- -..........((((....(-((((((((..-..(((((((((((((......)))))..)))).-)))).-...-..)))))))))......-))))......---- ( -34.30, z-score = -4.37, R) >droEre2.scaffold_4784 847518 90 - 25762168 --------GUAGAUCCUAGGAGGCGCUUUAAUUCACAGGUUUAGUGGAUC--CACACCACGAACA-CUGAC-UGU-ACUUAGUGCCAGAUAAA-UCGCGAUCAC---- --------...((((......((((((........(((((((.((((...--....)))))))).-)))..-...-....)))))).((....-))..))))..---- ( -22.83, z-score = -1.45, R) >droYak2.chr3L 827652 90 - 24197627 --------GUAGAUCGUAGCAGGCGCUUUAAUUCACAGGUUUAGUGGAUC--CACACCACGAACA-CUGAU-UGU-ACUUAGUGCCAGAUAAA-UUGUGAUCAC---- --------...(((((((((.((((((........(((((((.((((...--....)))))))).-)))..-...-....)))))).).....-))))))))..---- ( -21.53, z-score = -1.00, R) >droSec1.super_2 859758 90 - 7591821 --------GUAGACCCUAGAAGGCGCUUUAAUUCACAGGUUUAGUGGAUC--CACACCACGAACA-CUGAC-UGU-ACUUAGUGCCAGAUAAA-UCGUGAUCAC---- --------.............((((((........(((((((.((((...--....)))))))).-)))..-...-....)))))).(((...-.....)))..---- ( -19.93, z-score = -0.63, R) >droSim1.chr3L 486819 90 - 22553184 --------GUAGACCCUAGAAGGCGCUUUAAUUCACAGGUUUAGUGGAUA--CACACCACGAACA-CUGAC-UGU-ACUUAGUGCCAGAUAAA-UCGUGAUCAC---- --------.............((((((........(((((((.((((...--....)))))))).-)))..-...-....)))))).(((...-.....)))..---- ( -19.93, z-score = -0.77, R) >consensus ________CUAGAUCCUAGUUGGCGCUUUAAUUCACAGGGUUUGUGGAUCA_CACACCACGAACA_CUGAC_UGU_ACUUAGCGCCAGAUAAA_UUGUGAUCAC____ .....................((((((........(((.....((((.........))))......)))...........))))))...................... (-12.15 = -11.28 + -0.87)
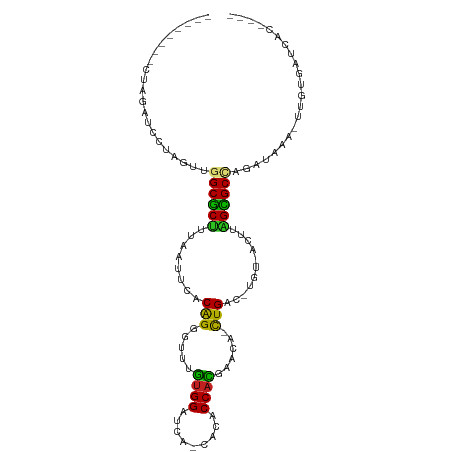
| Location | 831,946 – 832,036 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 59.40 |
| Shannon entropy | 0.88386 |
| G+C content | 0.44826 |
| Mean single sequence MFE | -23.47 |
| Consensus MFE | -8.85 |
| Energy contribution | -8.38 |
| Covariance contribution | -0.46 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.87 |
| SVM RNA-class probability | 0.995983 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 831946 90 - 24543557 ----CAUCAACUAGGUGGCGUGUAGACCCUAGAAGGCGCUUUAAUUCACAGGUUUAGUGGAUC--CACACCACGAACACUGAC-UGU-ACUUAGUGCC ----......(((((.(.(.....).))))))..((((((........(((((((.((((...--....)))))))).)))..-...-....)))))) ( -23.93, z-score = -0.60, R) >droVir3.scaffold_13049 16449196 96 + 25233164 -UAGCCGUACCAUCUGAAUAUCCUUCGUACCCUUGGCGCCUAAAUGUUCGGUGGCUAUGGCUAAUCCU-CCAUAGACACUGUUUUGUAACUGAGCGCU -(((((((((((.((((((((....(((.......))).....))))))))))).))))))))...((-(.....(((......)))....))).... ( -24.80, z-score = -1.43, R) >droWil1.scaffold_180698 9068707 89 - 11422946 ----CAUUCUCAUCUAUAUCUAUAUC-AAUAUUUUGCACUUCAAUUCUCAGAGACUGUGGAU--UCCA-CCAUAGUCUACUGACUGU-ACUUAGUGCU ----......................-........(((((...((..(((((((((((((..--....-))))))))).))))..))-....))))). ( -24.90, z-score = -4.63, R) >droGri2.scaffold_15110 20029156 91 + 24565398 ---UAAAUACAAAGCG-AUCUAUAUUUUUCACUUGGCGCUUUAAUUC-CAGUGUCUGUGGCUAAUAAAUCCAUGGACACUGCU-UGU-ACUUAGCGCU ---........(((.(-(..........)).)))((((((.((....-((((((((((((.........))))))))))))..-..)-)...)))))) ( -25.80, z-score = -3.53, R) >droMoj3.scaffold_6680 6556738 95 - 24764193 -UAUAUAUCUUAUCUAAAUUUCAUUCUAUUCUUUGGCGCUUUAAUUCACAGUGCAUAUGGAUAAUAGA-CCAUUGACAUUGUUAUGU-ACUUAGCGCU -.................................((((((.((....((((((((.((((........-)))))).))))))....)-)...)))))) ( -17.60, z-score = -1.46, R) >droPer1.super_29 463859 97 + 1099123 CAUCUUCCACUCUCUCCAUCUCUCUUUCCCUAUUAGCGCCUUAAUUCUCAGGGUUUGUGGAUUAACCCACCAUCGACACCUGUCUGU-ACUUGGCGCC ...................................(((((........(((((((.((((.........)))).))).)))).....-....))))). ( -21.63, z-score = -2.97, R) >dp4.chrXR_group8 4812076 97 + 9212921 CAUCUUCCACUCUCUCCAUCUCUCUUUCCCUAUUAGCGCCUUAAUUCUCAGGGUUUGUGGAUUAACCCACCAUCGACACCUGUCUGU-ACUUGGCGCC ...................................(((((........(((((((.((((.........)))).))).)))).....-....))))). ( -21.63, z-score = -2.97, R) >droAna3.scaffold_13337 11798714 89 - 23293914 ----GAUUGGUUUCAAAAUAC-UAGACAGUAGU-GGCGCUAUAG-ACACAGGUUCGGUGGAUCUUCCCACCCCGAACACUGUU-UAU-AAAUGGCGCC ----..(((((........))-)))........-((((((((..-..(((((((((((((......)))))..)))).)))).-...-..)))))))) ( -29.10, z-score = -2.91, R) >droEre2.scaffold_4784 847535 90 - 25762168 ----CAUCAACUAUGUGGCGUGUAGAUCCUAGGAGGCGCUUUAAUUCACAGGUUUAGUGGAUC--CACACCACGAACACUGAC-UGU-ACUUAGUGCC ----.........(((((.(((..(((((...(((.(.............).)))...)))))--))).)))))..((((((.-...-..)))))).. ( -22.52, z-score = -0.18, R) >droYak2.chr3L 827669 90 - 24197627 ----CAUCAACCAUGUGGCGUGUAGAUCGUAGCAGGCGCUUUAAUUCACAGGUUUAGUGGAUC--CACACCACGAACACUGAU-UGU-ACUUAGUGCC ----.........(((.(((.......))).)))((((((........(((((((.((((...--....)))))))).)))..-...-....)))))) ( -21.83, z-score = -0.08, R) >droSec1.super_2 859775 90 - 7591821 ----CAUAAACUAGGUGGCGUGUAGACCCUAGAAGGCGCUUUAAUUCACAGGUUUAGUGGAUC--CACACCACGAACACUGAC-UGU-ACUUAGUGCC ----......(((((.(.(.....).))))))..((((((........(((((((.((((...--....)))))))).)))..-...-....)))))) ( -23.93, z-score = -0.76, R) >droSim1.chr3L 486836 90 - 22553184 ----CAUUAACUAGGUGGCGUGUAGACCCUAGAAGGCGCUUUAAUUCACAGGUUUAGUGGAUA--CACACCACGAACACUGAC-UGU-ACUUAGUGCC ----......(((((.(.(.....).))))))..((((((........(((((((.((((...--....)))))))).)))..-...-....)))))) ( -23.93, z-score = -0.89, R) >consensus ____CAUUAACUACGUGAUGUGUAGAUCCUAGUUGGCGCUUUAAUUCACAGGGUUUGUGGAUC__CACACCACGAACACUGAU_UGU_ACUUAGCGCC ..................................((((((................((((.........))))..(((......))).....)))))) ( -8.85 = -8.38 + -0.46)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:54:28 2011