| Sequence ID | dm3.chr3L |
|---|---|
| Location | 826,750 – 826,840 |
| Length | 90 |
| Max. P | 0.870396 |

| Location | 826,750 – 826,840 |
|---|---|
| Length | 90 |
| Sequences | 11 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 62.45 |
| Shannon entropy | 0.70248 |
| G+C content | 0.33339 |
| Mean single sequence MFE | -12.85 |
| Consensus MFE | -6.10 |
| Energy contribution | -5.26 |
| Covariance contribution | -0.84 |
| Combinations/Pair | 1.67 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.870396 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
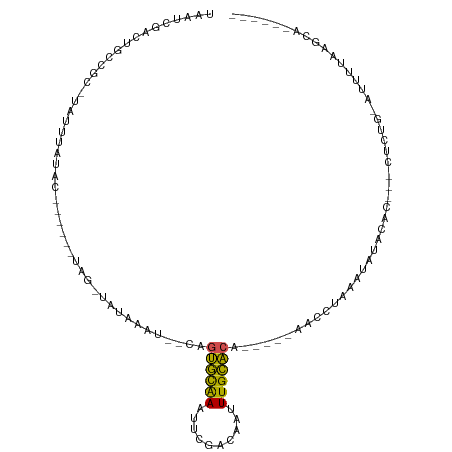
>dm3.chr3L 826750 90 - 24543557 UAAUCGACUGCCGCAUAUUUAAAC------UAG-CAUAUAAGUCAGUGCAAACUCGACAAAUUGCACA------ACCUAAAUAUAAAC---CCCUA-AUCAUAAGCA------ .....((((...((..........------..)-).....)))).((((((..........)))))).------..............---.....-..........------ ( -10.40, z-score = -1.06, R) >droSim1.chr3L 480078 86 - 22553184 UAAUCGACUGCCGCAUAUUUAAAC------UAG-CAUAUA----AGUGCAAACUCGACAAAUUGCACA------ACCUAAAUAUAAAC---CUCUA-AUCUUAAGCA------ .........((.((..........------..)-).....----.((((((..........)))))).------..............---.....-.......)).------ ( -8.10, z-score = -0.23, R) >droSec1.super_2 854600 86 - 7591821 UAAUCGACUGCCGCAUAUUUAAAC------UAG-CAUAUA----AGUGCAAACUCGACAAAUUGCACA------GCCUAAAUAUAAAC---CUCUA-AUCUUAAGCA------ .........((.((..........------..)-).....----.((((((..........)))))).------..............---.....-.......)).------ ( -8.10, z-score = 0.51, R) >droYak2.chr3L 822435 94 - 24197627 UAAUCGACUGCCGCAUAUUUACAC------UAG-CCUAAGU--CGGUGCAAAUUCGACAAUUUGCACAGCCACAACCUACAUAUAAAC---CACUA-AGUCUAAGCA------ ....(((((...((..........------..)-)...)))--))(((((((((....))))))))).....................---.....-..........------ ( -16.90, z-score = -1.80, R) >droEre2.scaffold_4784 842319 89 - 25762168 UAAUCGACUGCCGCAUAUUUAAAC------UAG-CAUAAGU-GCACUGCGAAUUCGACAAUUUGCACA------ACCUAAAUUUAAAC---CAUUA-AGUCUAAGCG------ .....((((........((((((.------..(-((....)-))..((((((((....))))))))..------.......)))))).---.....-))))......------ ( -12.74, z-score = -0.60, R) >droAna3.scaffold_13337 11793694 90 - 23293914 UAAUCGACUGCCGCCUAUUUAUAU------UAG-UCUAAGC--CACUGCGAAUGCCACAAUUUGCAAA-----AACCUAAAUAUUCAU---CCUAGCAUUUUAAGCA------ .....(((((..............------)))-)).....--...(((((((......)))))))..-----...............---....((.......)).------ ( -10.04, z-score = 0.13, R) >dp4.chrXR_group8 4807062 86 + 9212921 UAAACGACUGCC---UAUUUAUAC------UAG-UAUUUAAUUCAGUGCAAAUUCAAUGUUUUGCACA-----ACACCAAUUACCCAC---UUUAGCAUAGAAA--------- ........(((.---((((.....------.))-)).........(((((((........))))))).-----...............---....)))......--------- ( -10.00, z-score = -0.77, R) >droPer1.super_29 458839 86 + 1099123 UAAACGACUGCC---UAUUUAUAC------UAG-UAUUUAAUUCAGUGCAAAUUCAAUAUUUUGCACA-----ACACCAAUUACCCAC---AUUAGCAUAGAAA--------- ............---..(((((.(------(((-(...(((((..(((((((........))))))).-----.....))))).....---))))).)))))..--------- ( -10.80, z-score = -1.83, R) >droMoj3.scaffold_6680 6551587 104 - 24764193 UAAACGACUGCC---UAUUUAUAUUUAUACUAGAUUUAA-UUAUUGCAUAAAGUC-AACAUUUGUGCAU-CAUAUUUAAUUGCCACAC---UUGUGUAAUUUAAGCCACAAUA .........((.---((.((((((........((((.((-(...((((((((...-....)))))))).-...))).)))).......---..))))))..)).))....... ( -12.33, z-score = 0.52, R) >droVir3.scaffold_13049 16443954 97 + 25233164 UAAAUGACUGCC---UAUUUAUAC------UACAUUUAAUUUGUUGCACAAAUUCAAACAUUUGUGCAA-CAAAUUUAAUAGCCACACACGCUAUGUAAUUUAAGCC------ (((((.((....---.........------.......((((((((((((((((......))))))))))-))))))..(((((.......))))))).)))))....------ ( -23.30, z-score = -4.00, R) >droGri2.scaffold_15110 20023782 90 + 24565398 UAAAUGACUGCC---UAUUUAUAU------UAGAUGUAA-UUGUUGCACAAAUUGAAAUAUUUGUGCAG-CUUAUUUCAUAGCCACAC------UGUAAUUUAAGCA------ ..((..(.((((---((.......------)))..))).-)..))((((((((......)))))))).(-((((..(.((((.....)------))).)..))))).------ ( -18.60, z-score = -1.01, R) >consensus UAAUCGACUGCCGC_UAUUUAUAC______UAG_UAUAAAU__CAGUGCAAAUUCGACAAUUUGCACA_____AACCUAAAUAUACAC___CUCUG_AUUUUAAGCA______ .............................................((((((..........)))))).............................................. ( -6.10 = -5.26 + -0.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:54:26 2011