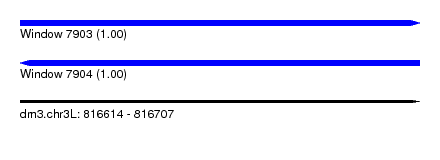
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 816,614 – 816,707 |
| Length | 93 |
| Max. P | 0.999772 |

| Location | 816,614 – 816,707 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 50.67 |
| Shannon entropy | 0.82504 |
| G+C content | 0.35286 |
| Mean single sequence MFE | -20.55 |
| Consensus MFE | -7.60 |
| Energy contribution | -9.22 |
| Covariance contribution | 1.63 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.36 |
| SVM RNA-class probability | 0.999772 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 816614 93 + 24543557 --------------AAAAAACUACACAGAAAAAAAA-----AGAAUUAAUUUUAA--CUUGCCGCAUAUCACAAACACUAUUUGCGGCGAAUUAAACAUCAUUGCCAUCACACU --------------......................-----.........(((((--.((((((((................)))))))).))))).................. ( -13.89, z-score = -3.33, R) >droEre2.scaffold_4784 832074 114 + 25762168 AAAAAAAGGAACAUAAAAGAUCGCGUUGAAUUAAACGAUUUAAAUUGGAUAUCAAAACUCGCCGCAGACCACAAACACUAUUUGCGGCGAGUUUUGCAUCAUUGCCAACACACC .......((..((....((((((..(......)..))))))....))(((..((((((((((((((((............)))))))))))))))).)))....))........ ( -31.40, z-score = -4.62, R) >droYak2.chr3L 811957 103 + 24197627 AAAUAAUGGAACAGAAAAAAAAG-----AAAUAAA------AAAUGGGAUAUCAAAACUCGCCGCAAACCAUAACCACUAUUUGCGGCGAAUUUAACAUCAUUGCCAACACACU ......(((....((........-----.......------...(((....)))(((.((((((((((............)))))))))).)))....))....)))....... ( -17.90, z-score = -2.83, R) >droMoj3.scaffold_6496 17934691 107 + 26866924 AAACUCGUAUUCUGAUAAAAUGAAGUCUUAUUUUGA-----UUAGCAAGCAAUACAGCUCGCACCACUGUGCGAAUAAAUACCGAGCUACACUCAACAC--UCAUCUGCAUAUU ......(((((((((((((((((....))))))).)-----)))).....))))).((((((((....)))))).........(((.....))).....--......))..... ( -19.00, z-score = -1.19, R) >consensus AAA_AA_GGAACAGAAAAAAUAACGU_GAAAUAAAA_____AAAUUGAAUAUCAAAACUCGCCGCAAACCACAAACACUAUUUGCGGCGAAUUUAACAUCAUUGCCAACACACU ..........................................................(((((((((((..........)))))))))))........................ ( -7.60 = -9.22 + 1.63)

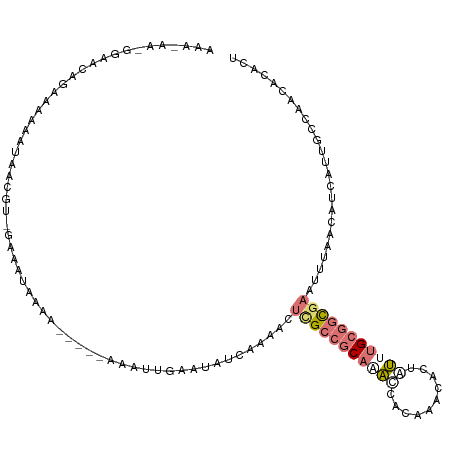
| Location | 816,614 – 816,707 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 50.67 |
| Shannon entropy | 0.82504 |
| G+C content | 0.35286 |
| Mean single sequence MFE | -25.59 |
| Consensus MFE | -12.41 |
| Energy contribution | -12.60 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.65 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.39 |
| SVM RNA-class probability | 0.998541 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 816614 93 - 24543557 AGUGUGAUGGCAAUGAUGUUUAAUUCGCCGCAAAUAGUGUUUGUGAUAUGCGGCAAG--UUAAAAUUAAUUCU-----UUUUUUUUCUGUGUAGUUUUUU-------------- .............((((.(((((((.((((((.((..........)).)))))).))--))))))))).....-----......................-------------- ( -14.10, z-score = 0.63, R) >droEre2.scaffold_4784 832074 114 - 25762168 GGUGUGUUGGCAAUGAUGCAAAACUCGCCGCAAAUAGUGUUUGUGGUCUGCGGCGAGUUUUGAUAUCCAAUUUAAAUCGUUUAAUUCAACGCGAUCUUUUAUGUUCCUUUUUUU ((((((((((.((((((.((((((((((((((.((..........)).)))))))))))))).............))))))....))))))).))).................. ( -33.44, z-score = -3.09, R) >droYak2.chr3L 811957 103 - 24197627 AGUGUGUUGGCAAUGAUGUUAAAUUCGCCGCAAAUAGUGGUUAUGGUUUGCGGCGAGUUUUGAUAUCCCAUUU------UUUAUUU-----CUUUUUUUUCUGUUCCAUUAUUU .......(((....(((((((((((((((((((((.((....)).)))))))))))).))))))))))))...------.......-----....................... ( -27.20, z-score = -3.59, R) >droMoj3.scaffold_6496 17934691 107 - 26866924 AAUAUGCAGAUGA--GUGUUGAGUGUAGCUCGGUAUUUAUUCGCACAGUGGUGCGAGCUGUAUUGCUUGCUAA-----UCAAAAUAAGACUUCAUUUUAUCAGAAUACGAGUUU ....(((..((((--(((..(((.....)))..)))))))..)))...((((((((((......))))))..)-----))).....((((((.((((.....))))..)))))) ( -27.60, z-score = -0.97, R) >consensus AGUGUGUUGGCAAUGAUGUUAAAUUCGCCGCAAAUAGUGUUUGUGGUAUGCGGCGAGUUUUAAUAUCCAAUUU_____UUUUAAUUC_ACGCAAUUUUUUCUGUUCC_UU_UUU ..(((....)))..((((((((((((((((((.((..........)).))))))))))).)))))))............................................... (-12.41 = -12.60 + 0.19)

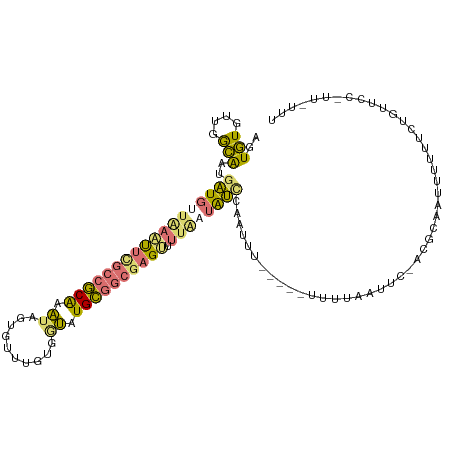
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:54:25 2011