
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,085,263 – 4,085,329 |
| Length | 66 |
| Max. P | 0.667323 |

| Location | 4,085,263 – 4,085,329 |
|---|---|
| Length | 66 |
| Sequences | 13 |
| Columns | 76 |
| Reading direction | forward |
| Mean pairwise identity | 83.07 |
| Shannon entropy | 0.36251 |
| G+C content | 0.39937 |
| Mean single sequence MFE | -13.72 |
| Consensus MFE | -7.64 |
| Energy contribution | -7.65 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.667323 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
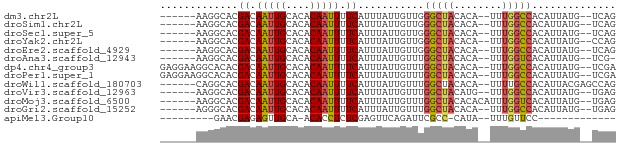
>dm3.chr2L 4085263 66 + 23011544 ------AAGGCACGACAAUUGCACACAAUUUUCAUUUAUUGUUGGGCUACACA--UUUGGCCACAUUAUG--UCAG ------..(((((((((((..................)))))))(((((....--..)))))......))--)).. ( -14.47, z-score = -2.23, R) >droSim1.chr2L 4040467 66 + 22036055 ------AAGGCACGACAAUUGCACACAAUUUUCAUUUAUUGUUGGGCUACACA--UUUGGCCACAUUAUG--UCAG ------..(((((((((((..................)))))))(((((....--..)))))......))--)).. ( -14.47, z-score = -2.23, R) >droSec1.super_5 2184419 66 + 5866729 ------AAGGCACGACAAUUGCACACAAUUUUCAUUUAUUGUUGGGCUACACA--UUUGGCCACAUUAUG--UCAG ------..(((((((((((..................)))))))(((((....--..)))))......))--)).. ( -14.47, z-score = -2.23, R) >droYak2.chr2L 4102423 66 + 22324452 ------AAGGCACGACAAUUGCACACAAUUUUCAUUUAUUGUUGGGCUACACA--UUUGGCCACAUUAUG--CCAG ------..(((((((((((..................)))))))(((((....--..)))))......))--)).. ( -17.17, z-score = -3.01, R) >droEre2.scaffold_4929 4143340 66 + 26641161 ------AAGGCACGACAAUUGCACACAAUUUUCAUUUAUUGUUGGGCUACACA--UUUGGCCACAUUAUG--UCAG ------..(((((((((((..................)))))))(((((....--..)))))......))--)).. ( -14.47, z-score = -2.23, R) >droAna3.scaffold_12943 652304 65 - 5039921 ------AAGGCACGACAAUUGCACACAAUUUUCAUUUAUUGUUUGGCUACACA--UUUGGUCACAUUAUG--UCG- ------......(((((.......(((((........))))).((((((....--..)))))).....))--)))- ( -11.20, z-score = -1.44, R) >dp4.chr4_group3 9833522 72 - 11692001 GAGGAAGGCACACGACAAUUGCACACAAUUUUCAUUUAUUGUUUGGCUACACA--UUUGGCCACAUUAUG--UCGA ............(((((.......(((((........))))).((((((....--..)))))).....))--))). ( -15.00, z-score = -1.91, R) >droPer1.super_1 6951647 72 - 10282868 GAGGAAGGCACACGACAAUUGCACACAAUUUUCAUUUAUUGUUUGGCUACACA--UUUGGCCACAUUAUG--UCGA ............(((((.......(((((........))))).((((((....--..)))))).....))--))). ( -15.00, z-score = -1.91, R) >droWil1.scaffold_180703 2079272 68 + 3946847 ------CAGGCACGACAAUUGCACACAAUUUUCAUUUAUUGUUUGGCUACACA--UUUUGCCACAUUACGAGCCAG ------..(((..((.(((((....))))).)).....((((.((((......--....))))....))))))).. ( -14.90, z-score = -3.30, R) >droVir3.scaffold_12963 11054664 66 - 20206255 ------AAGGCACGACAAUUGCACACAAUUUUCAUUUAUUGUUUGGCUACAUG--UUUGGCCACAUUAUG--UGAG ------....(((((.(((((....))))).))..........((((((....--..))))))......)--)).. ( -12.80, z-score = -0.60, R) >droMoj3.scaffold_6500 2178634 68 + 32352404 ------AAGGCACGACAAUUGCACACAAUUUUCAUUUAUUGUUUGGCUACACACAUUUGGUCACAUUAUG--UGAG ------..(((..((((((..................))))))..)))...(((((.((....))..)))--)).. ( -10.57, z-score = -0.42, R) >droGri2.scaffold_15252 12667186 66 + 17193109 ------AGGGCACGACAAUUGCACACAAUUUUCAUUUAUUGUUUGGCUACACA--UUUGGCCACAUUAUG--UGAG ------....(((((.(((((....))))).))..........((((((....--..))))))......)--)).. ( -12.80, z-score = -1.19, R) >apiMel3.Group10 6020629 50 - 11440700 ---------GAACGAGAGUUGCA-ACACCUCUCGAGUUCAGAUUCGCC-CAUA--UUUGUUCC------------- ---------(((((((((.((..-.)).)))))((((....))))...-....--...)))).------------- ( -11.00, z-score = -1.97, R) >consensus ______AAGGCACGACAAUUGCACACAAUUUUCAUUUAUUGUUUGGCUACACA__UUUGGCCACAUUAUG__UCAG .............((.(((((....))))).))...........(((((........))))).............. ( -7.64 = -7.65 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:15:18 2011