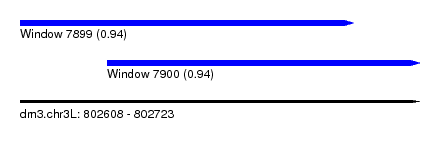
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 802,608 – 802,723 |
| Length | 115 |
| Max. P | 0.944720 |

| Location | 802,608 – 802,704 |
|---|---|
| Length | 96 |
| Sequences | 8 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 70.72 |
| Shannon entropy | 0.56420 |
| G+C content | 0.53354 |
| Mean single sequence MFE | -18.55 |
| Consensus MFE | -10.51 |
| Energy contribution | -10.90 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.944720 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
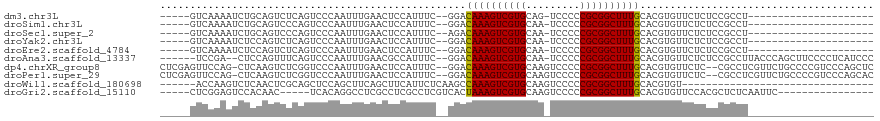
>dm3.chr3L 802608 96 + 24543557 ---------------AGCCAUGUAGCCCAGUCCAGUCCACGUCAAAAUCUGCAGUCUCAGUCCCAAUUUGAACUCCAUUUC--GGACAAAGUCGUGCAG-UCCCCCGCGGCUUU ---------------((((..((.((........))..))((......((((((.((..((((.((..((.....)).)).--))))..)).).)))))-......)))))).. ( -18.40, z-score = -1.27, R) >droSim1.chr3L 453976 96 + 22553184 ---------------AGCCAUGUAGCCCAGUCCAGUCCACGUCAAAAUCUGCAGUCCCAGUCCCAAUUUGAACUCCAUUUC--GGACAAAGUCGUGCAA-UCCCCCGCGGCUUU ---------------..............((((........(((((..(((......)))......)))))..........--))))(((((((((...-.....))))))))) ( -16.67, z-score = -0.98, R) >droSec1.super_2 823800 96 + 7591821 ---------------AGCCAUGUAGCCCAGUCCAGUCCACGUCAAAAUCUGCAGUCCCAGUCCCAAUUUGAACUCCAUUUC--AGACAAAGUCGUGCAA-UCCCCCGCGGCUUU ---------------.........................(((.....(((......)))........((((......)))--))))(((((((((...-.....))))))))) ( -13.90, z-score = -0.81, R) >droYak2.chr3L 797188 103 + 24197627 --------AGCCAUAUACUCCAUACCCCAGUCCAGUCCACGUCAAAAUCUCCAGUCUCAGUCCCAAUUUGAACUCCAUUUC--GGACAAAGUCGUGCAA-UCCCCCGCGGCUUU --------.....................((((.(....)............(((.((((.......))))))).......--))))(((((((((...-.....))))))))) ( -16.10, z-score = -2.12, R) >droEre2.scaffold_4784 817693 100 + 25762168 ----------CCAUAUACCCCA-GUCCAAGUCCAGUCCACGUCAAAAUCUCCAGUCUCAGUCCCAAUUUGAACUCCAUUUC--GGACAAAGUCGUGCAA-UCCCCCGCGGCUUU ----------............-((((..((.......))............(((.((((.......))))))).......--))))(((((((((...-.....))))))))) ( -16.20, z-score = -2.66, R) >droAna3.scaffold_13337 11768353 88 + 23293914 ----------------------ACCCGAAGCUAUAACCCAA-CUCCGACUCCAGUUUCAGUCCCAAUUUGAACGCCAUUUC--GGACAAAGUCGUGCAA-UCCCCCGCGGCUUU ----------------------....((((((.........-..........)))))).((((.((..((.....)).)).--))))(((((((((...-.....))))))))) ( -17.61, z-score = -1.59, R) >dp4.chrXR_group8 4781896 112 - 9212921 AUCGGCGUCGGGCAUCGGCGUCGGCGUCGGCUCCGUCUCGAGUUCCAGCUCAAGUCUCGGUCCCAAUUUGAACUCCAUUUC--GGACAAAGUCGUGCAAGUCCCCCGCGGCUUU .(((((((((.....))))))))).(((((..(((.((.((((....)))).))...)))..))....((((......)))--))))(((((((((.........))))))))) ( -37.10, z-score = -0.56, R) >droWil1.scaffold_180698 9030300 87 + 11422946 ---------------------------AACCAAAACCCCCAACCAAGUCUCAACUCGCAGCUCCAGCUUCAGCUUCAUUCUCAAGCCAAAGUCGUGCAAGUCCCCCGCGGCUUU ---------------------------.............................(.(((....))).).((((.......)))).(((((((((.........))))))))) ( -12.40, z-score = -1.31, R) >consensus _______________AGCCAUGUACCCCAGUCCAGUCCACGUCAAAAUCUCCAGUCUCAGUCCCAAUUUGAACUCCAUUUC__GGACAAAGUCGUGCAA_UCCCCCGCGGCUUU ...........................................................((((.....((.....))......))))(((((((((.........))))))))) (-10.51 = -10.90 + 0.39)

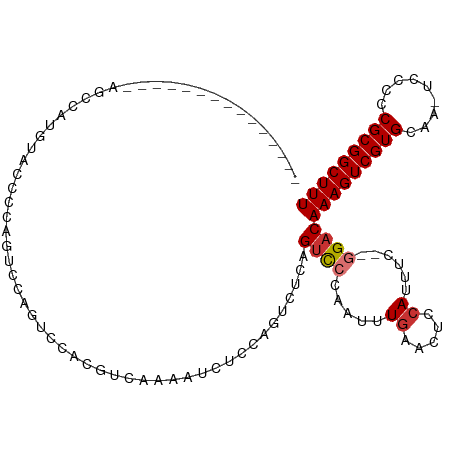
| Location | 802,633 – 802,723 |
|---|---|
| Length | 90 |
| Sequences | 10 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 70.57 |
| Shannon entropy | 0.57001 |
| G+C content | 0.55313 |
| Mean single sequence MFE | -20.34 |
| Consensus MFE | -11.38 |
| Energy contribution | -11.29 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.938852 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 802633 90 + 24543557 -----GUCAAAAUCUGCAGUCUCAGUCCCAAUUUGAACUCCAUUUC--GGACAAAGUCGUGCAG-UCCCCCGCGGCUUUGCACGUGUUCUCUCCGCCU--------------------- -----........((((((.((..((((.((..((.....)).)).--))))..)).).)))))-......((((....((....)).....))))..--------------------- ( -18.90, z-score = -1.25, R) >droSim1.chr3L 454001 90 + 22553184 -----GUCAAAAUCUGCAGUCCCAGUCCCAAUUUGAACUCCAUUUC--GGACAAAGUCGUGCAA-UCCCCCGCGGCUUUGCACGUGUUCUCUCCGCCU--------------------- -----.(((((..(((......)))......))))).........(--(((......(((((((-..((....))..))))))).......))))...--------------------- ( -18.22, z-score = -1.43, R) >droSec1.super_2 823825 90 + 7591821 -----GUCAAAAUCUGCAGUCCCAGUCCCAAUUUGAACUCCAUUUC--AGACAAAGUCGUGCAA-UCCCCCGCGGCUUUGCACGUGUUCUCUCCGCCU--------------------- -----..........((..............((((((......)))--)))((((((((((...-.....))))))))))..............))..--------------------- ( -16.30, z-score = -1.47, R) >droYak2.chr3L 797220 90 + 24197627 -----GUCAAAAUCUCCAGUCUCAGUCCCAAUUUGAACUCCAUUUC--GGACAAAGUCGUGCAA-UCCCCCGCGGCUUUGCACGUGUUCUCUCCGCCU--------------------- -----............(((.((((.......)))))))......(--(((......(((((((-..((....))..))))))).......))))...--------------------- ( -17.82, z-score = -1.92, R) >droEre2.scaffold_4784 817722 90 + 25762168 -----GUCAAAAUCUCCAGUCUCAGUCCCAAUUUGAACUCCAUUUC--GGACAAAGUCGUGCAA-UCCCCCGCGGCUUUGCACGUGUUCUCUCCGCCU--------------------- -----............(((.((((.......)))))))......(--(((......(((((((-..((....))..))))))).......))))...--------------------- ( -17.82, z-score = -1.92, R) >droAna3.scaffold_13337 11768373 108 + 23293914 ------UCCGA--CUCCAGUUUCAGUCCCAAUUUGAACGCCAUUUC--GGACAAAGUCGUGCAA-UCCCCCGCGGCUUUGCACGUGUUCUCUCCGCCUUACCCAGCUUCCCCUCAUCCC ------...((--((........)))).......((((((......--(..((((((((((...-.....))))))))))..))))))).....((........))............. ( -21.00, z-score = -1.67, R) >dp4.chrXR_group8 4781932 114 - 9212921 CUCGAGUUCCAG-CUCAAGUCUCGGUCCCAAUUUGAACUCCAUUUC--GGACAAAGUCGUGCAAGUCCCCCGCGGCUUUGCACGUGUUCUC--CGCCUCGUUCUGCCCCGUCCCAGCUC ...((((....)-))).......((..(......((((.......(--(((.((...((((((((.((.....)).))))))))..)).))--))....))))......)..))..... ( -27.00, z-score = -1.12, R) >droPer1.super_29 433465 114 - 1099123 CUCGAGUUCCAG-CUCAAGUCUCGGUCCCAAUUUGAACUCCAUUUC--GGACAAAGUCGUGCAAGUCCCCCGCGGCUUUGCACGUGUUCUC--CGCCUCGUUCUGCCCCGUCCCAGCAC ...((((....)-))).......((..(......((((.......(--(((.((...((((((((.((.....)).))))))))..)).))--))....))))......)..))..... ( -27.00, z-score = -0.98, R) >droWil1.scaffold_180698 9030314 81 + 11422946 ------ACCAAGUCUCAACUCGCAGCUCCAGCUUCAGCUUCAUUCUCAAGCCAAAGUCGUGCAAGUCCCCCGCGGCUUUGCACGUGU-------------------------------- ------..............(((.......((((..((((.......))))..)))).(((((((.((.....)).)))))))))).-------------------------------- ( -17.30, z-score = -1.15, R) >droGri2.scaffold_15110 19996788 93 - 24565398 -----CUCGGAGUCCACAAC-----UCACAGGCCUCGCCUCGCCUCGUCACUAAAGUCGUGCAAGUCCCCCGCGGCUUUGCACGUGUUCCACGCUCUCAAUUC---------------- -----...((((..(((...-----..((((((........)))).))..........(((((((.((.....)).)))))))))))))).............---------------- ( -22.00, z-score = -1.02, R) >consensus _____GUCAAAAUCUCCAGUCUCAGUCCCAAUUUGAACUCCAUUUC__GGACAAAGUCGUGCAA_UCCCCCGCGGCUUUGCACGUGUUCUCUCCGCCU_____________________ ...................................................((((((((((.........))))))))))....................................... (-11.38 = -11.29 + -0.09)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:54:22 2011