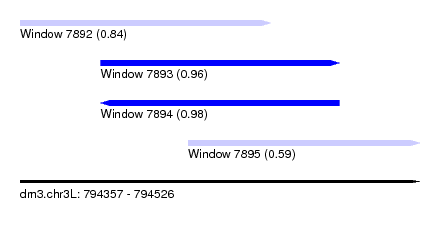
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 794,357 – 794,526 |
| Length | 169 |
| Max. P | 0.984080 |

| Location | 794,357 – 794,463 |
|---|---|
| Length | 106 |
| Sequences | 10 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 80.82 |
| Shannon entropy | 0.40988 |
| G+C content | 0.43819 |
| Mean single sequence MFE | -34.51 |
| Consensus MFE | -17.97 |
| Energy contribution | -18.34 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.838525 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 794357 106 + 24543557 ------UGUGUGCAUAAAUUUGUGUGCAUGAUCGGCAGCAAUAAUUUUAUUUGUUUACGAUUAUGUUCGCUGGCCAA-CAGGCAAUGGCCAGUAAGCCAGUCAGUUGGCUUCU ------.....(((((....)))))(((((((((..(((((.........)))))..)))))))))..((((((((.-.......))))))))(((((((....))))))).. ( -36.50, z-score = -2.69, R) >droAna3.scaffold_13337 11757125 106 + 23293914 ------GGUGGGCAUAAAUUUGUGUGCAUGAUCGGCAGCAAUAAUUUUAUUUGUUUACGAUUAUGUUCGCUGGCCAA-CAGGCAAUGGCCAGUCAGCCGGCCAAUGGUCAGCC ------(((.(((..........(.(((((((((..(((((.........)))))..))))))))).)((((((((.-.......))))))))..))).)))........... ( -40.10, z-score = -2.77, R) >droEre2.scaffold_4784 809320 106 + 25762168 ------UGUGUGCAUAAAUUUGUGUGCAUGAUCGGCAGCAAUAAUUUUAUUUGUUUACGAUUAUGUUCGCUGGCCAA-CAGGCAAUGGCCAGUAAGCCAGUCAGUUGGCUCCU ------.....(((((....)))))(((((((((..(((((.........)))))..)))))))))..((((((((.-.......)))))))).((((((....))))))... ( -35.70, z-score = -2.30, R) >droYak2.chr3L 788855 106 + 24197627 ------UGUGUGCAUAAAUUUGUGUGCAUGAUCGGCAGCAAUAAUUUUAUUUGUUUACGAUUAUGUUCGCUGGCCAA-CAGGCAAUGGCCAGUAAGCCAGUCAGUUGGCUUCU ------.....(((((....)))))(((((((((..(((((.........)))))..)))))))))..((((((((.-.......))))))))(((((((....))))))).. ( -36.50, z-score = -2.69, R) >droSec1.super_2 815559 106 + 7591821 ------UGUGUGCAUAAAUUUGUGUGCAUGAUCGGCAGCAAUAAUUUUAUUUGUUUACGAUUAUGUUCCCUGGCCAA-CAGGCAAUGGCCAGUAAGCCAGUCAGUUGGCUCCU ------.....(((((....)))))(((((((((..(((((.........)))))..)))))))))...(((((((.-.......)))))))..((((((....))))))... ( -33.60, z-score = -1.87, R) >droSim1.chr3L 445226 106 + 22553184 ------UGUGUGCAUAAAUUUGUGUGCAUGAUCGGCAGCAAUAAUUUUAUUUGUUUACGAUUAUGUUCGCUGGCCAA-CAGGCAAUGGCCAGUAAGCCAGUCAGUUGGCUCCU ------.....(((((....)))))(((((((((..(((((.........)))))..)))))))))..((((((((.-.......)))))))).((((((....))))))... ( -35.70, z-score = -2.30, R) >droPer1.super_29 423936 111 - 1099123 UGUGUUUCUUAGCAUAAAUUUGUGUGCAUGAUCGGCAGCAAUAAUUUUAUUUGUUUACGAUUAUGUUCGCUGGCCAACAGGCUCUUGUACUGCCGGGGACCCGAGGCGCCU-- .(((((((...(((((....)))))(((((((((..(((((.........)))))..)))))))))((.(((((..((((....))))...))))).))...)))))))..-- ( -34.70, z-score = -1.40, R) >dp4.chrXR_group8 4772464 111 - 9212921 UGUGUUUCUUAGCAUAAAUUUGUGUGCAUGAUCGGCAGCAAUAAUUUUAUUUGUUUACGAUUAUGUUCGCUGGCCAACAGGCUCUUGUACUGCCGGGGACCCGAGGCGCCU-- .(((((((...(((((....)))))(((((((((..(((((.........)))))..)))))))))((.(((((..((((....))))...))))).))...)))))))..-- ( -34.70, z-score = -1.40, R) >droWil1.scaffold_180698 9018408 105 + 11422946 ------GGUGUGCAUAAAUUUGUGUGCAUGAUCGGCAGCAAUAAUUUUAUUUGUUUACGAUUAUGUUCGCUGGCCAA-CAGCCAAUAGCCAG-AUGCUGCUGACCAGGUUUGU ------.(((..((((....))))..)))(.(((((((((((((((.((......)).)))))).....(((((...-.........)))))-.))))))))).)........ ( -31.80, z-score = -1.17, R) >droVir3.scaffold_13049 16405067 87 - 25233164 -------GUGUGCAUAAAUUUGUGUGCAUGAUCGGCAGCAAUAAUUUUAUUUGUUUACGAUUAUGUUCGCUGCCCAAACAGGCAACAAGCGAUC------------------- -------(((..((((....))))..)))((((((((((.((((((.((......)).))))))....))))))......(....)....))))------------------- ( -25.80, z-score = -2.02, R) >consensus ______UGUGUGCAUAAAUUUGUGUGCAUGAUCGGCAGCAAUAAUUUUAUUUGUUUACGAUUAUGUUCGCUGGCCAA_CAGGCAAUGGCCAGUAAGCCAGUCAGUUGGCUUCU ...........(((((....)))))(((((((((..(((((.........)))))..)))))))))..(((((...............))))).................... (-17.97 = -18.34 + 0.37)

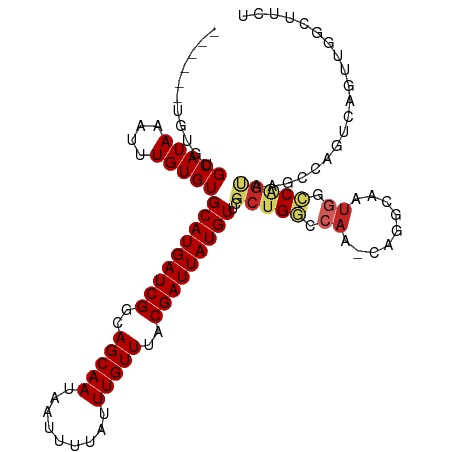
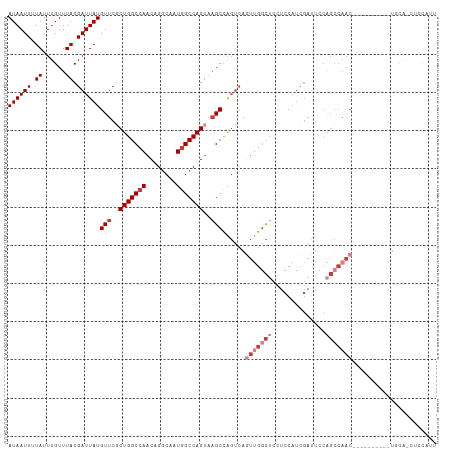
| Location | 794,391 – 794,492 |
|---|---|
| Length | 101 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 77.51 |
| Shannon entropy | 0.42760 |
| G+C content | 0.46384 |
| Mean single sequence MFE | -28.27 |
| Consensus MFE | -17.55 |
| Energy contribution | -19.41 |
| Covariance contribution | 1.86 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.964058 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 794391 101 + 24543557 AUAAUUUUAUUUGUUUACGAUUAUGUUCGCUGGCCAACAGGCAAUGGCCAGUAAGCCAGUCAGUUGGCUUCUCCAUCGAUUCCAGCCAACUCCGGCCAACU----------- ((((((.((......)).))))))(((..(((((((........)))))))..)))..(((((((((((((......))....))))))))..))).....----------- ( -29.20, z-score = -2.57, R) >droAna3.scaffold_13337 11757159 100 + 23293914 AUAAUUUUAUUUGUUUACGAUUAUGUUCGCUGGCCAACAGGCAAUGGCCAGUCAGCCGGCCAAUGGUCAGCCGCAGCCAACCAGGCCAAC----------UCCA--UUCAUU ((((((.((......)).))))))(((.((((((((........)))))))).))).((((..((((..((....))..))))))))...----------....--...... ( -30.50, z-score = -2.05, R) >droEre2.scaffold_4784 809354 112 + 25762168 AUAAUUUUAUUUGUUUACGAUUAUGUUCGCUGGCCAACAGGCAAUGGCCAGUAAGCCAGUCAGUUGGCUCCUCCAUCGAUUCCAGCCAACUGCGGCCAACUCCAGCUCCAUU ((((((.((......)).))))))....((((((((........))))))))..(((.(.(((((((((..((....))....)))))))))))))................ ( -32.60, z-score = -2.44, R) >droYak2.chr3L 788889 112 + 24197627 AUAAUUUUAUUUGUUUACGAUUAUGUUCGCUGGCCAACAGGCAAUGGCCAGUAAGCCAGUCAGUUGGCUUCUCCAUCGAUUCCAGCCAACGCCGGCCAACUGCAACUCCAUU ((((((.((......)).))))))(((.((((((.....(((..((((......))))))).(((((((((......))....))))))))))))).)))............ ( -31.20, z-score = -2.01, R) >droSec1.super_2 815593 102 + 7591821 AUAAUUUUAUUUGUUUACGAUUAUGUUCCCUGGCCAACAGGCAAUGGCCAGUAAGCCAGUCAGUUGGCUCCUCCAUCGAUUCCAGCCAAC----------UGCAGCUCCAUU ((((((.((......)).))))))(((..(((((((........)))))))..)))..(.(((((((((..((....))....)))))))----------)))......... ( -28.70, z-score = -2.73, R) >droSim1.chr3L 445260 102 + 22553184 AUAAUUUUAUUUGUUUACGAUUAUGUUCGCUGGCCAACAGGCAAUGGCCAGUAAGCCAGUCAGUUGGCUCCUCCAUCGAUUCCAGCCAAC----------UGCAGCUCCAUU ((((((.((......)).))))))(((..(((((((........)))))))..)))..(.(((((((((..((....))....)))))))----------)))......... ( -30.60, z-score = -2.95, R) >droWil1.scaffold_180698 9018442 82 + 11422946 AUAAUUUUAUUUGUUUACGAUUAUGUUCGCUGGCCAACAGCCAAUAGCCAG-AUGCUGCUGACCAGGUUUGUUCCUUGUUUGC----------------------------- ((((((.((......)).))))))((...(((((............)))))-..)).((.(((.(((......))).))).))----------------------------- ( -15.10, z-score = 0.37, R) >consensus AUAAUUUUAUUUGUUUACGAUUAUGUUCGCUGGCCAACAGGCAAUGGCCAGUAAGCCAGUCAGUUGGCUCCUCCAUCGAUUCCAGCCAAC__________UGCA_CUCCAUU ((((((.((......)).))))))(((..(((((((........)))))))..)))......(((((((..............)))))))...................... (-17.55 = -19.41 + 1.86)

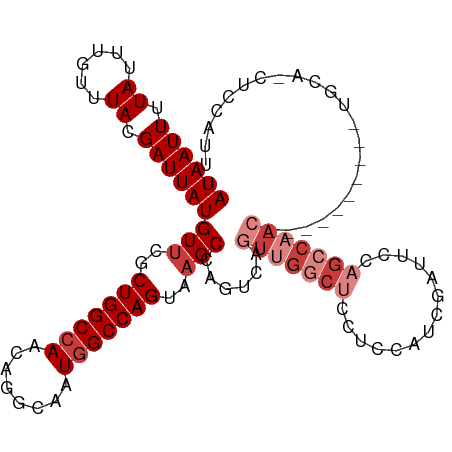
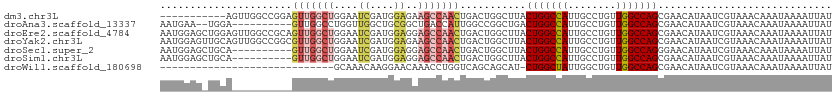
| Location | 794,391 – 794,492 |
|---|---|
| Length | 101 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 77.51 |
| Shannon entropy | 0.42760 |
| G+C content | 0.46384 |
| Mean single sequence MFE | -34.73 |
| Consensus MFE | -16.70 |
| Energy contribution | -17.89 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.984080 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 794391 101 - 24543557 -----------AGUUGGCCGGAGUUGGCUGGAAUCGAUGGAGAAGCCAACUGACUGGCUUACUGGCCAUUGCCUGUUGGCCAGCGAACAUAAUCGUAAACAAAUAAAAUUAU -----------(((.((((((((((((((....((....))..))))))))..)))))).)))(((((........))))).((((......))))................ ( -36.10, z-score = -3.46, R) >droAna3.scaffold_13337 11757159 100 - 23293914 AAUGAA--UGGA----------GUUGGCCUGGUUGGCUGCGGCUGACCAUUGGCCGGCUGACUGGCCAUUGCCUGUUGGCCAGCGAACAUAAUCGUAAACAAAUAAAAUUAU .(((((--((.(----------(((((((((((..((....))..))))..))))))))(.(((((((........))))))))...)))..))))................ ( -39.90, z-score = -3.51, R) >droEre2.scaffold_4784 809354 112 - 25762168 AAUGGAGCUGGAGUUGGCCGCAGUUGGCUGGAAUCGAUGGAGGAGCCAACUGACUGGCUUACUGGCCAUUGCCUGUUGGCCAGCGAACAUAAUCGUAAACAAAUAAAAUUAU .(((..((((((((.((((.(((((((((....((....))..)))))))))...)))).)))(((....)))......)))))...)))...................... ( -38.00, z-score = -2.28, R) >droYak2.chr3L 788889 112 - 24197627 AAUGGAGUUGCAGUUGGCCGGCGUUGGCUGGAAUCGAUGGAGAAGCCAACUGACUGGCUUACUGGCCAUUGCCUGUUGGCCAGCGAACAUAAUCGUAAACAAAUAAAAUUAU ..((...((((.(((((((((((((((((....((....))..)))))))....(((((....)))))......))))))))))((......)))))).))........... ( -39.20, z-score = -2.73, R) >droSec1.super_2 815593 102 - 7591821 AAUGGAGCUGCA----------GUUGGCUGGAAUCGAUGGAGGAGCCAACUGACUGGCUUACUGGCCAUUGCCUGUUGGCCAGGGAACAUAAUCGUAAACAAAUAAAAUUAU .((((((((.((----------(((((((....((....))..)))))))))...))))).(((((((........)))))))....)))...................... ( -35.00, z-score = -3.04, R) >droSim1.chr3L 445260 102 - 22553184 AAUGGAGCUGCA----------GUUGGCUGGAAUCGAUGGAGGAGCCAACUGACUGGCUUACUGGCCAUUGCCUGUUGGCCAGCGAACAUAAUCGUAAACAAAUAAAAUUAU .((((((((.((----------(((((((....((....))..)))))))))...))))).(((((((........)))))))....)))...................... ( -34.10, z-score = -2.83, R) >droWil1.scaffold_180698 9018442 82 - 11422946 -----------------------------GCAAACAAGGAACAAACCUGGUCAGCAGCAU-CUGGCUAUUGGCUGUUGGCCAGCGAACAUAAUCGUAAACAAAUAAAAUUAU -----------------------------.......(((......)))((((((((((..-..........)))))))))).((((......))))................ ( -20.80, z-score = -2.07, R) >consensus AAUGGAG_UGCA__________GUUGGCUGGAAUCGAUGGAGAAGCCAACUGACUGGCUUACUGGCCAUUGCCUGUUGGCCAGCGAACAUAAUCGUAAACAAAUAAAAUUAU ......................(((((((....((....))..)))))))...........(((((((........)))))))............................. (-16.70 = -17.89 + 1.19)
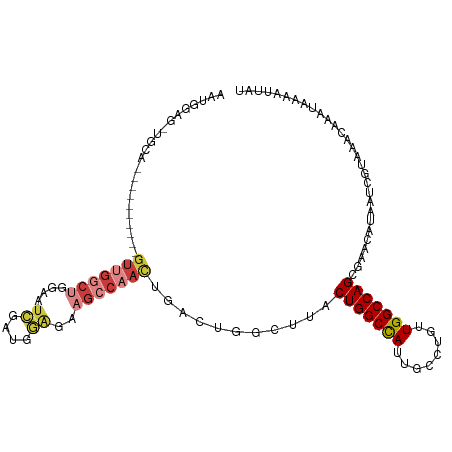
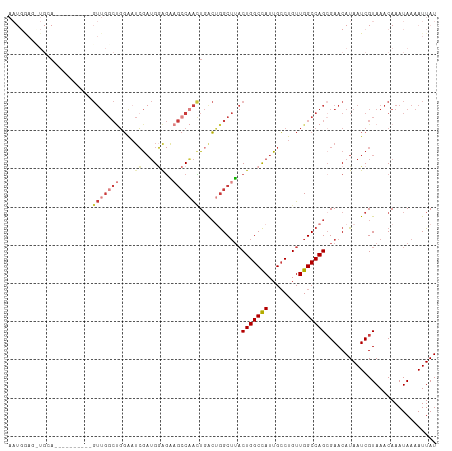
| Location | 794,428 – 794,526 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 72.80 |
| Shannon entropy | 0.50864 |
| G+C content | 0.58857 |
| Mean single sequence MFE | -27.65 |
| Consensus MFE | -14.14 |
| Energy contribution | -15.85 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.590622 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 794428 98 + 24543557 CAGGCAAUGGCCAGUAAGCCAGUCAGUUGGCUUCUCCAUCGAUUCCAGCCAACUCCGGCCAACUGCGG-------------AGUCGCGUCACUCAUACGCCCCGACGACAG---- ..(((..((((......))))...((((((((((......))....))))))))...)))..(((((.-------------.(..((((.......))))..)..)).)))---- ( -30.70, z-score = -1.69, R) >droAna3.scaffold_13337 11757196 102 + 23293914 CAGGCAAUGGCCAGUCAGCCGGCCAAUGGUCAGCCGCAGCCAACCAGGCCAACUCCA------------UUCAUU-GCCGGAGUCGCGUCACUCAUACGCCCCGAUGACAGCAGC ..(((((((((......)))((((..((((..((....))..)))))))).......------------...)))-)))(..(((((((.......))))...)))..)...... ( -31.80, z-score = -0.63, R) >droEre2.scaffold_4784 809391 111 + 25762168 CAGGCAAUGGCCAGUAAGCCAGUCAGUUGGCUCCUCCAUCGAUUCCAGCCAACUGCGGCCAACUCCAGCUCCAUUCAGCAAAGUCGCGUCACUCAUACGCCCCGACGACAG---- ..(((..((((......))))(.(((((((((..((....))....)))))))))).))).......(((......)))...(((((((.......))))...))).....---- ( -31.20, z-score = -2.28, R) >droYak2.chr3L 788926 111 + 24197627 CAGGCAAUGGCCAGUAAGCCAGUCAGUUGGCUUCUCCAUCGAUUCCAGCCAACGCCGGCCAACUGCAACUCCAUUCAGCAAAGUCGCGUCACUCAUACGCCCCGACGACAG---- ...(((((((.((((..(((.((..(((((((((......))....))))))))).)))..)))).....)))))..))...(((((((.......))))...))).....---- ( -28.40, z-score = -1.31, R) >droSec1.super_2 815630 101 + 7591821 CAGGCAAUGGCCAGUAAGCCAGUCAGUUGGCUCCUCCAUCGAUUCCAGCCAACUGC----------AGCUCCAUUCAGCAGAGUCGCGUCACUCAUACGCCCCGACGACAG---- ..(((..((((......))))(.(((((((((..((....))....))))))))))----------.(((......))).((((......))))....)))..........---- ( -27.70, z-score = -1.66, R) >droSim1.chr3L 445297 101 + 22553184 CAGGCAAUGGCCAGUAAGCCAGUCAGUUGGCUCCUCCAUCGAUUCCAGCCAACUGC----------AGCUCCAUUCAGCAGAGUCGCGUCACUCAUACGCCCCGACGACAG---- ..(((..((((......))))(.(((((((((..((....))....))))))))))----------.(((......))).((((......))))....)))..........---- ( -27.70, z-score = -1.66, R) >droWil1.scaffold_180698 9018479 75 + 11422946 CAGCCAAUAGCCAG-AUGCUGCUGACCAGGUUUGUUCCUUGUUUGC-----------------------------------AGUCGCGUCACUCAUACGCCCCGACGACAG---- ..............-..(((((.(((.(((......))).))).))-----------------------------------))).(((((.............)))).)..---- ( -16.02, z-score = 0.36, R) >consensus CAGGCAAUGGCCAGUAAGCCAGUCAGUUGGCUCCUCCAUCGAUUCCAGCCAACUCC__________AGCUCCAUUCAGCAGAGUCGCGUCACUCAUACGCCCCGACGACAG____ .......((((......))))(((((((((((..............))))))))............................(((((((.......))))...))))))...... (-14.14 = -15.85 + 1.72)

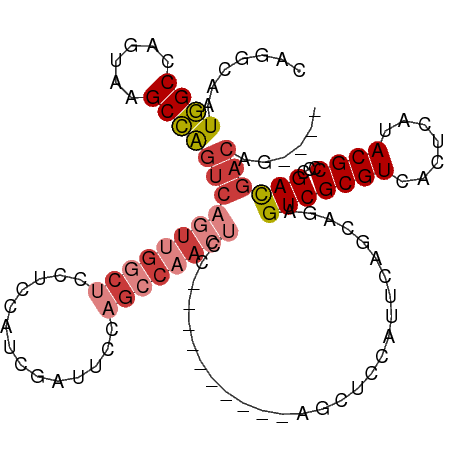
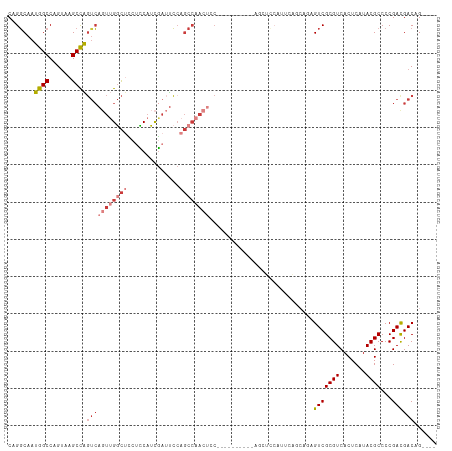
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:54:18 2011