| Sequence ID | dm3.chr3L |
|---|---|
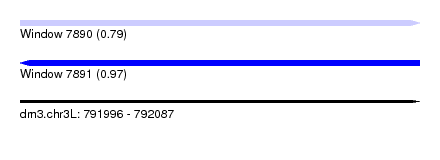
| Location | 791,996 – 792,087 |
| Length | 91 |
| Max. P | 0.970919 |

| Location | 791,996 – 792,087 |
|---|---|
| Length | 91 |
| Sequences | 10 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 85.19 |
| Shannon entropy | 0.27714 |
| G+C content | 0.36468 |
| Mean single sequence MFE | -17.97 |
| Consensus MFE | -12.18 |
| Energy contribution | -11.86 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.790407 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 791996 91 + 24543557 -------------------GUCUCGUCUCGUCGUCUCGAAACAGACAAUAAAACAUCAUUAAAAU--CCAUCGGCGUUGUAACGCGCCUUGCAAAUAUUUUAAUUGUUUUUC -------------------((((.((.(((......))).))))))...((((((..((((((((--.....(((((......)))))........)))))))))))))).. ( -19.52, z-score = -3.09, R) >droSec1.super_2 812759 91 + 7591821 -------------------GUCUCGUCUCGUCGUCUCAAAACAGACAAUAAAACAUCAUUAAAAU--CCAUCGGCGUUGUAACACGCCUUGCAAAUAUUUUAAUUGUUUUUC -------------------.............((((......))))...((((((..((((((((--.....(((((......)))))........)))))))))))))).. ( -14.52, z-score = -2.44, R) >droSim1.chr3L 442863 91 + 22553184 -------------------GUCUCGUCUCGUCGUCUCGAAACAGACAAUAAAACAUCAUUAAAAG--CCAUCGGCGUUGUAACACGCCUUGCAAAUAUUUUAAUUGUUUUUC -------------------((((.((.(((......))).))))))...((((((..(((((((.--.....(((((......))))).........))))))))))))).. ( -19.06, z-score = -2.85, R) >droYak2.chr3L 786447 91 + 24197627 -------------------GUCUUGUCUCGUCGUCUCGAAACAGACAAUAAAACAUCAUUAAAAU--CCAUCGGCGUUGUAACACGCCUUGCAAAUAUUUUAAUUGUUUUUC -------------------((((.((.(((......))).))))))...((((((..((((((((--.....(((((......)))))........)))))))))))))).. ( -19.22, z-score = -3.31, R) >droEre2.scaffold_4784 805167 91 + 25762168 -------------------GUCUCGUCUCGUCGUCUCGAAACAGACAAUAAAACAUCAUUAAAAU--CCAUCGGCGUUGUAACACGCCUUGCAAAUAUUUUAAUUGUUUUUC -------------------((((.((.(((......))).))))))...((((((..((((((((--.....(((((......)))))........)))))))))))))).. ( -19.22, z-score = -3.48, R) >droAna3.scaffold_13337 11754820 93 + 23293914 ---------------GAGGCGUUCGUCUGGCCGUC--GAAACAGACAAUAAAACAUCAUUAAAAA--CCAACAGCGUUGUAACACGCCUUGCAAAUAUUUUAAUUGUUUUUC ---------------(((((((..(((((......--....)))))...................--.((((...))))....)))))))...................... ( -17.30, z-score = -0.70, R) >dp4.chrXR_group8 4769032 90 - 9212921 ------------------GUGUUCGUCUCGCCGUC--GAAACAGACAAUAAAAUAUCAUUAAAAA--UCAACGGCGUUGUAACACGCCUUGCAAAUAUUUUAAUUGUUUUUC ------------------((.((((.(.....).)--)))))((((((((((((((.........--.(((.(((((......))))))))...))))))).)))))))... ( -15.82, z-score = -1.04, R) >droPer1.super_29 420524 90 - 1099123 ------------------GUGUUCGUCUCGCCGUC--GAAACAGACAAUAAAAUAUCAUUAAAAA--UCAACGGCGUUGUAACACGCCUUGCAAAUAUUUUAAUUGUUUUUC ------------------((.((((.(.....).)--)))))((((((((((((((.........--.(((.(((((......))))))))...))))))).)))))))... ( -15.82, z-score = -1.04, R) >droWil1.scaffold_180698 9013293 93 + 11422946 ---------------GUCUUGUUUGUCUGGCCAUC--GAAACAGACAAUAAAACAUCAUUAAAAC--UCAACGGCGUUGUAAUACGCCUUGCAAAUAUUUUAAUUGUUUUUC ---------------.......(((((((......--....))))))).((((((..(((((((.--.(((.(((((......))))))))......))))))))))))).. ( -20.00, z-score = -2.55, R) >droGri2.scaffold_15110 19983792 110 - 24565398 GUUGGUUUCUUUGCUUGUGUUUUUGUCUAGCCGUC--GAAGCAGACAAUAAAACAUCAUUAAAAAAAUCAGAGGCGUUGUAACAUGCUCUGCAAAUAUUUUAAUUGUUUUUC (((((..(.(((((..((((((((((((.((....--...)))))))..))))))).............((((.(((......)))))))))))).)..)))))........ ( -19.20, z-score = 0.15, R) >consensus ___________________GUCUCGUCUCGCCGUC__GAAACAGACAAUAAAACAUCAUUAAAAA__CCAUCGGCGUUGUAACACGCCUUGCAAAUAUUUUAAUUGUUUUUC ........................((((..............))))...((((((..(((((((........(((((......))))).........))))))))))))).. (-12.18 = -11.86 + -0.32)

| Location | 791,996 – 792,087 |
|---|---|
| Length | 91 |
| Sequences | 10 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 85.19 |
| Shannon entropy | 0.27714 |
| G+C content | 0.36468 |
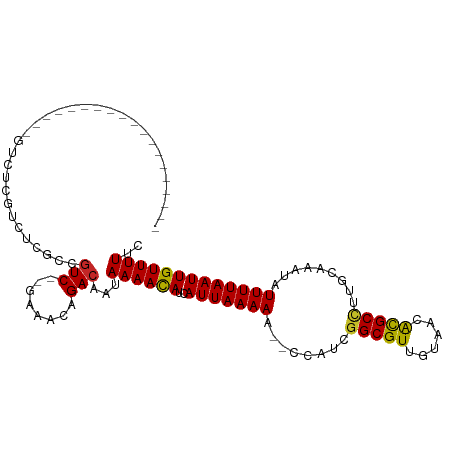
| Mean single sequence MFE | -22.70 |
| Consensus MFE | -17.91 |
| Energy contribution | -17.78 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.970919 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 791996 91 - 24543557 GAAAAACAAUUAAAAUAUUUGCAAGGCGCGUUACAACGCCGAUGG--AUUUUAAUGAUGUUUUAUUGUCUGUUUCGAGACGACGAGACGAGAC------------------- ..((((((((((((((........((((........)))).....--))))))))..))))))...((((((((((......)))))).))))------------------- ( -22.22, z-score = -2.11, R) >droSec1.super_2 812759 91 - 7591821 GAAAAACAAUUAAAAUAUUUGCAAGGCGUGUUACAACGCCGAUGG--AUUUUAAUGAUGUUUUAUUGUCUGUUUUGAGACGACGAGACGAGAC------------------- ........((((((((........(((((......))))).....--))))))))..(((((..((((((......))))))..)))))....------------------- ( -23.22, z-score = -2.99, R) >droSim1.chr3L 442863 91 - 22553184 GAAAAACAAUUAAAAUAUUUGCAAGGCGUGUUACAACGCCGAUGG--CUUUUAAUGAUGUUUUAUUGUCUGUUUCGAGACGACGAGACGAGAC------------------- ..(((((((((((((.....((..(((((......)))))....)--))))))))..))))))...((((((((((......)))))).))))------------------- ( -24.70, z-score = -2.81, R) >droYak2.chr3L 786447 91 - 24197627 GAAAAACAAUUAAAAUAUUUGCAAGGCGUGUUACAACGCCGAUGG--AUUUUAAUGAUGUUUUAUUGUCUGUUUCGAGACGACGAGACAAGAC------------------- ..((((((((((((((........(((((......))))).....--))))))))..))))))...((((((((((......)))))).))))------------------- ( -24.92, z-score = -3.39, R) >droEre2.scaffold_4784 805167 91 - 25762168 GAAAAACAAUUAAAAUAUUUGCAAGGCGUGUUACAACGCCGAUGG--AUUUUAAUGAUGUUUUAUUGUCUGUUUCGAGACGACGAGACGAGAC------------------- ..((((((((((((((........(((((......))))).....--))))))))..))))))...((((((((((......)))))).))))------------------- ( -24.92, z-score = -3.18, R) >droAna3.scaffold_13337 11754820 93 - 23293914 GAAAAACAAUUAAAAUAUUUGCAAGGCGUGUUACAACGCUGUUGG--UUUUUAAUGAUGUUUUAUUGUCUGUUUC--GACGGCCAGACGAACGCCUC--------------- .......................((((((........((((((((--..(.((((((....))))))...)..))--)))))).......)))))).--------------- ( -21.06, z-score = -0.78, R) >dp4.chrXR_group8 4769032 90 + 9212921 GAAAAACAAUUAAAAUAUUUGCAAGGCGUGUUACAACGCCGUUGA--UUUUUAAUGAUAUUUUAUUGUCUGUUUC--GACGGCGAGACGAACAC------------------ ..........(((((((((..((((((((......))))).))).--........))))))))).(((.((((((--(....))))))).))).------------------ ( -22.00, z-score = -2.11, R) >droPer1.super_29 420524 90 + 1099123 GAAAAACAAUUAAAAUAUUUGCAAGGCGUGUUACAACGCCGUUGA--UUUUUAAUGAUAUUUUAUUGUCUGUUUC--GACGGCGAGACGAACAC------------------ ..........(((((((((..((((((((......))))).))).--........))))))))).(((.((((((--(....))))))).))).------------------ ( -22.00, z-score = -2.11, R) >droWil1.scaffold_180698 9013293 93 - 11422946 GAAAAACAAUUAAAAUAUUUGCAAGGCGUAUUACAACGCCGUUGA--GUUUUAAUGAUGUUUUAUUGUCUGUUUC--GAUGGCCAGACAAACAAGAC--------------- ..((((((((((((((.....((((((((......))))).))).--))))))))..)))))).(((((((....--......))))))).......--------------- ( -22.10, z-score = -2.27, R) >droGri2.scaffold_15110 19983792 110 + 24565398 GAAAAACAAUUAAAAUAUUUGCAGAGCAUGUUACAACGCCUCUGAUUUUUUUAAUGAUGUUUUAUUGUCUGCUUC--GACGGCUAGACAAAAACACAAGCAAAGAAACCAAC ..(((((((((((((......(((((..(((....))).)))))....)))))))..)))))).(((((((((..--...))).))))))...................... ( -19.90, z-score = -1.27, R) >consensus GAAAAACAAUUAAAAUAUUUGCAAGGCGUGUUACAACGCCGAUGG__AUUUUAAUGAUGUUUUAUUGUCUGUUUC__GACGACGAGACGAAAC___________________ ..(((((((((((((.........(((((......)))))........)))))))..)))))).(((((((((.......))).))))))...................... (-17.91 = -17.78 + -0.13)
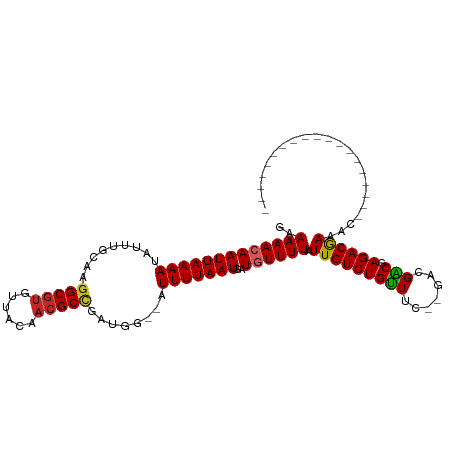
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:54:15 2011