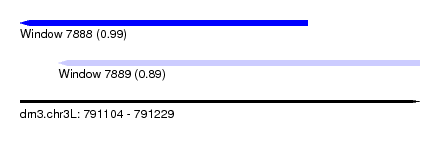
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 791,104 – 791,229 |
| Length | 125 |
| Max. P | 0.991204 |

| Location | 791,104 – 791,194 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 78.56 |
| Shannon entropy | 0.41131 |
| G+C content | 0.52261 |
| Mean single sequence MFE | -34.27 |
| Consensus MFE | -23.86 |
| Energy contribution | -23.40 |
| Covariance contribution | -0.46 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.991204 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
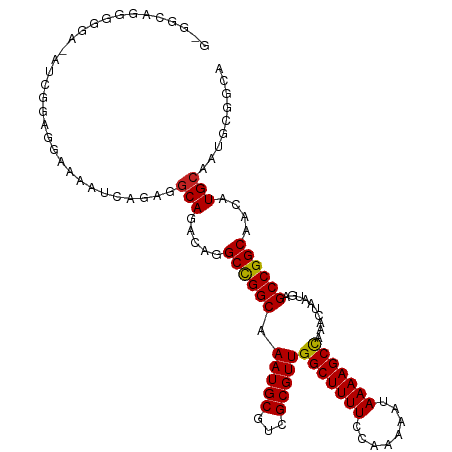
>dm3.chr3L 791104 90 - 24543557 ---ACAGGCCGGCAAAUGCGUCGCGUUGGCUUUUCCAAAAAUAAAAGCCAAACUAAUGAGCCGGCAACAUGCGAUGCAACAACU-----GCGACGCAC-------------------- ---....((((((.(((((...)))))(((((((........)))))))..........))))))....((((.((((.....)-----))).)))).-------------------- ( -33.20, z-score = -3.27, R) >droSec1.super_2 811961 90 - 7591821 ---ACAGGCCGGCAAAUGCGUCGCGUUGGCUUUUCCAAAAAUAAAAGCCAAACUAAUGAGCCGGCAACAUGCGAUGCAACAACU-----GCGACGCAC-------------------- ---....((((((.(((((...)))))(((((((........)))))))..........))))))....((((.((((.....)-----))).)))).-------------------- ( -33.20, z-score = -3.27, R) >droSim1.chr3L 442030 90 - 22553184 ---ACAGGCCGGCAAAUGCGUCGCGUUGGCUUUUCCAAAAAUAAAAGCCAAACUAAUGAGCCGGCAACAUGCGAUGCAACAACU-----GCGACGCAC-------------------- ---....((((((.(((((...)))))(((((((........)))))))..........))))))....((((.((((.....)-----))).)))).-------------------- ( -33.20, z-score = -3.27, R) >droYak2.chr3L 785559 90 - 24197627 ---ACAGGCCGGCAAAUGCGUCGCGUUGGCUUUUCCAAAAAUAAAAGCCAAACUAAUGAGCCGGCAACAUGCGAUGCAACAACU-----GCGACGCAC-------------------- ---....((((((.(((((...)))))(((((((........)))))))..........))))))....((((.((((.....)-----))).)))).-------------------- ( -33.20, z-score = -3.27, R) >droEre2.scaffold_4784 804261 90 - 25762168 ---ACAGGCCGGCAAAUGCGUCGCGUUGGCUUUUCCAAAAAUAAAAGCCAAACUAAUGAGCCGGCAACAUGCGAUGCAACAACU-----GCGACGCAC-------------------- ---....((((((.(((((...)))))(((((((........)))))))..........))))))....((((.((((.....)-----))).)))).-------------------- ( -33.20, z-score = -3.27, R) >droAna3.scaffold_13337 11753674 111 - 23293914 ---ACAGGCCGGCAAAUGCGUCGCGUUGGCUUUUCCAAAAAUAAAAGCCAAACUAAUGAGCCGGCAACAUGCAAUGCGGCAGCAGC--AGCAGC--AACAACAACUGUUGCAACGCAC ---....((((((.(((((...)))))(((((((........)))))))..........))))))....(((..(((.((....))--.)))((--((((.....))))))...))). ( -40.00, z-score = -2.31, R) >dp4.chrXR_group8 4767865 107 + 9212921 ---ACAGGCCGGCGAAUGCGUCGCGUUGGCUUUUCCAAAAAUAAAAGCCAAACUAAUGAGCCGGCAACAUGCAAUGCGGC--------AGCAGCAGAGCAGCCACAACUGAGACGCAC ---.(((((((((.....(((....(((((((((........)))))))))....))).)))))).........((.(((--------.((......)).))).)).)))........ ( -36.00, z-score = -1.52, R) >droPer1.super_29 419309 115 + 1099123 ---ACAGGCCGGCGAAUGCGUCGCGUUGGCUUUUCCAAAAAUAAAAGCCAAACUAAUGAGCCGGCAACAUGCAAUGCGGCAGCAGCAGAGCAGCAGAGCAGCCACAACUGAGACGCAC ---.(((((((((.....(((....(((((((((........)))))))))....))).))))))....(((..(((.((.........)).)))..))).......)))........ ( -36.70, z-score = -0.81, R) >droWil1.scaffold_180698 9011805 102 - 11422946 ---ACAGGCCGGCAAAUGCGUCGCGUUGGCUUUUCCAAAAAUAAAAGCUAAACUAAUGAGCCGGCAACAUGCAAUGCGGC--------AGC-----AGCAACUGCAACUGAGACGCAC ---....((((((.(((((...)))))(((((((........)))))))..........)))))).........((((.(--------((.-----.((....))..)))...)))). ( -32.20, z-score = -1.00, R) >droVir3.scaffold_13049 16401799 86 + 25233164 ---ACAGGCUGGC-AAUGCGUCGCGUUGGCUUUUCCAAAAAUAAAAGCCAAACUAAUGAGCCGGCAACAUGCAAUGCGGCAGCGACG-CAC--------------------------- ---..........-..((((((((.(((((((((........)))))))))........((((.((........)))))).))))))-)).--------------------------- ( -32.70, z-score = -2.60, R) >droMoj3.scaffold_6680 6510684 93 - 24764193 --GACAGGCUGGC-AAUGCGUCGCGUUGGCUUUUCCAAAAAUAAAAGCCAAACUAAUGAGCCGGCAACAUGCAAUGCGGCAGCGACG--ACGACGCAC-------------------- --...........-..((((((((((((((((((........)))))))..........((((.((........))))))...))))--.))))))).-------------------- ( -31.60, z-score = -1.75, R) >droGri2.scaffold_15110 19983050 96 + 24565398 ACAGUCGGCCGGCAAAUGCGUCGCGUUGGCUUUUCCAAAAAUAAAAGCCAAACUAAUGAGCCGGCAACAUGCAAUGCGGCAGUCGCU--GCGACGCAC-------------------- ...(((.((((((.(((((...)))))(((((((........)))))))..........))))))..........(((((....)))--)))))....-------------------- ( -36.10, z-score = -2.03, R) >consensus ___ACAGGCCGGCAAAUGCGUCGCGUUGGCUUUUCCAAAAAUAAAAGCCAAACUAAUGAGCCGGCAACAUGCAAUGCGGCAACU_____GCGACGCAC____________________ .......((((((.(((((...)))))(((((((........)))))))..........))))))....(((...)))........................................ (-23.86 = -23.40 + -0.46)

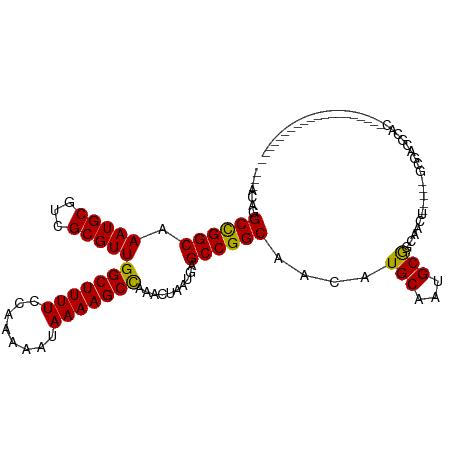
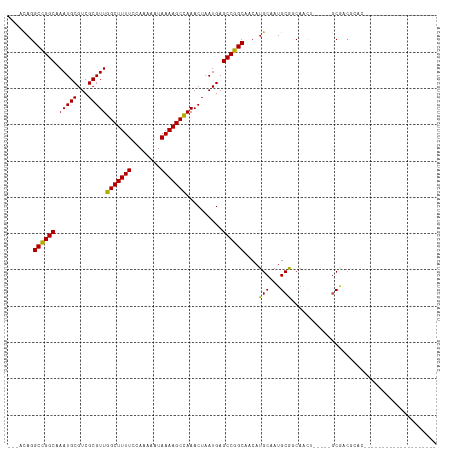
| Location | 791,116 – 791,229 |
|---|---|
| Length | 113 |
| Sequences | 11 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 79.57 |
| Shannon entropy | 0.44453 |
| G+C content | 0.51080 |
| Mean single sequence MFE | -31.51 |
| Consensus MFE | -27.09 |
| Energy contribution | -26.95 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.891294 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
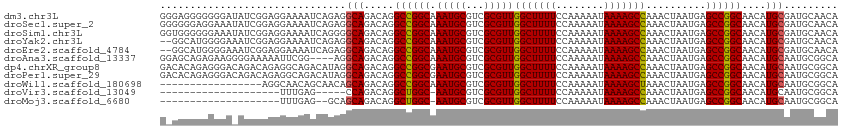
>dm3.chr3L 791116 113 - 24543557 GGGAGGGGGGGAUAUCGGAGGAAAAUCAGAGGCAGACAGGCCGGCAAAUGCGUCGCGUUGGCUUUUCCAAAAAUAAAAGCCAAACUAAUGAGCCGGCAACAUGCGAUGCAACA .......(..(.((((.((......))....(((.....((((((.(((((...)))))(((((((........)))))))..........))))))....))))))))..). ( -32.10, z-score = -0.83, R) >droSec1.super_2 811973 113 - 7591821 GGGGGGAGGAAAUAUCGGAGGAAAAUCAGAGGCAGACAGGCCGGCAAAUGCGUCGCGUUGGCUUUUCCAAAAAUAAAAGCCAAACUAAUGAGCCGGCAACAUGCGAUGCAACA ............((((.((......))....(((.....((((((.(((((...)))))(((((((........)))))))..........))))))....)))))))..... ( -30.40, z-score = -0.60, R) >droSim1.chr3L 442042 113 - 22553184 GGUGGGGGGAAAUAUCGGAGGAAAAUCAGGGGCAGACAGGCCGGCAAAUGCGUCGCGUUGGCUUUUCCAAAAAUAAAAGCCAAACUAAUGAGCCGGCAACAUGCGAUGCAACA ((((........))))............(..(((..((.((((((.(((((...)))))(((((((........)))))))..........))))))....))...)))..). ( -30.90, z-score = -0.25, R) >droYak2.chr3L 785571 111 - 24197627 --GGCAUGGGGAAAUCGGAGGAAAAUCAGAGGCAGACAGGCCGGCAAAUGCGUCGCGUUGGCUUUUCCAAAAAUAAAAGCCAAACUAAUGAGCCGGCAACAUGCGAUGCAACA --(((.((..(...((.((......)).))..)...)).)))......((((((((((((((((((........))))))))............(....)))))))))))... ( -33.60, z-score = -1.35, R) >droEre2.scaffold_4784 804273 111 - 25762168 --GGCAUGGGGAAAUCGGAGGAAAAUCAGAGGCAGACAGGCCGGCAAAUGCGUCGCGUUGGCUUUUCCAAAAAUAAAAGCCAAACUAAUGAGCCGGCAACAUGCGAUGCAACA --(((.((..(...((.((......)).))..)...)).)))......((((((((((((((((((........))))))))............(....)))))))))))... ( -33.60, z-score = -1.35, R) >droAna3.scaffold_13337 11753707 109 - 23293914 GGAGCAGAGAAGGGGAAAAAUUCGG----AGGCAGACAGGCCGGCAAAUGCGUCGCGUUGGCUUUUCCAAAAAUAAAAGCCAAACUAAUGAGCCGGCAACAUGCAAUGCGGCA ...(((..(((.........)))..----..(((.....((((((.(((((...)))))(((((((........)))))))..........))))))....)))..))).... ( -31.70, z-score = -0.58, R) >dp4.chrXR_group8 4767894 113 + 9212921 GACACAGAGGGACAGACAGAGGCAGACAUAGGCAGACAGGCCGGCGAAUGCGUCGCGUUGGCUUUUCCAAAAAUAAAAGCCAAACUAAUGAGCCGGCAACAUGCAAUGCGGCA .....................((.(.(((..(((.....((((((.....(((....(((((((((........)))))))))....))).))))))....))).)))).)). ( -32.30, z-score = -1.23, R) >droPer1.super_29 419346 113 + 1099123 GACACAGAGGGACAGACAGAGGCAGACAUAGGCAGACAGGCCGGCGAAUGCGUCGCGUUGGCUUUUCCAAAAAUAAAAGCCAAACUAAUGAGCCGGCAACAUGCAAUGCGGCA .....................((.(.(((..(((.....((((((.....(((....(((((((((........)))))))))....))).))))))....))).)))).)). ( -32.30, z-score = -1.23, R) >droWil1.scaffold_180698 9011829 96 - 11422946 -----------------AGGCAACAGCAACAGCAGACAGGCCGGCAAAUGCGUCGCGUUGGCUUUUCCAAAAAUAAAAGCUAAACUAAUGAGCCGGCAACAUGCAAUGCGGCA -----------------..((....(((...(((.....((((((.(((((...)))))(((((((........)))))))..........))))))....)))..))).)). ( -30.70, z-score = -1.28, R) >droVir3.scaffold_13049 16401808 87 + 25233164 --------------------UUUGAG-----CCAGACAGGCUGGC-AAUGCGUCGCGUUGGCUUUUCCAAAAAUAAAAGCCAAACUAAUGAGCCGGCAACAUGCAAUGCGGCA --------------------.....(-----((((.....)))))-.....(((((((((((((((........)))))))..........((.(....)..)))))))))). ( -29.90, z-score = -1.57, R) >droMoj3.scaffold_6680 6510699 90 - 24764193 --------------------UUUGAG--GCAGCAGACAGGCUGGC-AAUGCGUCGCGUUGGCUUUUCCAAAAAUAAAAGCCAAACUAAUGAGCCGGCAACAUGCAAUGCGGCA --------------------......--((.(((..((.((((((-....(((....(((((((((........)))))))))....))).))))))....))...))).)). ( -29.10, z-score = -0.73, R) >consensus G_GGCAGGGGGA_AUCGGAGGAAAAUCAGAGGCAGACAGGCCGGCAAAUGCGUCGCGUUGGCUUUUCCAAAAAUAAAAGCCAAACUAAUGAGCCGGCAACAUGCAAUGCGGCA ...............................(((.....((((((.(((((...)))))(((((((........)))))))..........))))))....)))......... (-27.09 = -26.95 + -0.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:54:13 2011