| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,073,766 – 4,073,856 |
| Length | 90 |
| Max. P | 0.708864 |

| Location | 4,073,766 – 4,073,856 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 81.34 |
| Shannon entropy | 0.39803 |
| G+C content | 0.39782 |
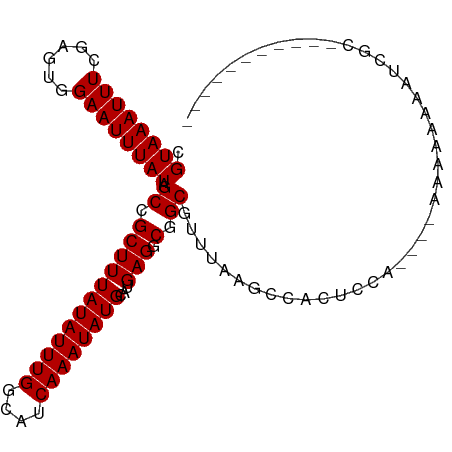
| Mean single sequence MFE | -23.62 |
| Consensus MFE | -11.48 |
| Energy contribution | -11.48 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
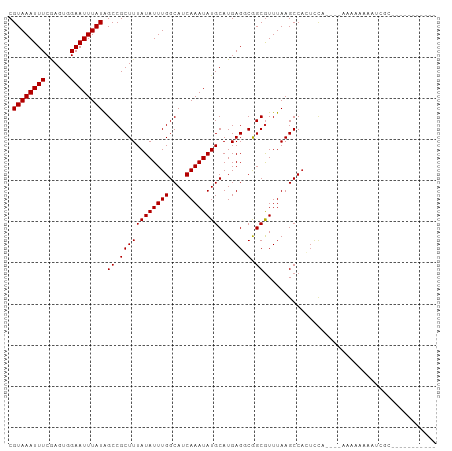
| Mean z-score | -2.05 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.708864 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
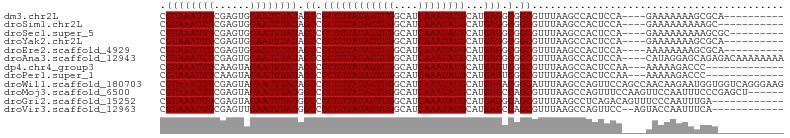
>dm3.chr2L 4073766 90 - 23011544 CGUAAAUUUCGAGUGGAAUUUAUAGCCGCUUUAUAUUUGGCAUCAAAUAUGCAUGAGGCGGCGUUUAAGCCACUCCA----GAAAAAAAGCGCA---------- (((...((((((((((........((((((((((.....((((.....)))))))))))))).......))))))..----))))....)))..---------- ( -28.56, z-score = -3.04, R) >droSim1.chr2L 4027740 89 - 22036055 CGUAAAUUUCGAGUGGAAUUUAUAGCCGCUUUAUAUUUGGCAUCAAAUAUGCAUGAGGCGGCGUUUAAGCCACUCCA----GAAAAAAAAAGC----------- ......((((((((((........((((((((((.....((((.....)))))))))))))).......))))))..----))))........----------- ( -27.96, z-score = -3.67, R) >droSec1.super_5 2172796 91 - 5866729 CGUAAAUUUCGAGUGGAAUUUAUAGCCGCUUUAUAUUUGGCAUCAAAUAUGCAUGAGGCGGCGUUUAAGCCACUCCA----GAAAAAAAAAGCGC--------- ......((((((((((........((((((((((.....((((.....)))))))))))))).......))))))..----))))..........--------- ( -27.96, z-score = -3.08, R) >droYak2.chr2L 4090540 90 - 22324452 CGUAAAUUUCGAGUGGAAUUUAUAGCCGCUUUAUAUUUGGCAUCAAAUAUGCAUGAGGCGGCGUUUAAGCCACUCCA----GAAAAAAAGCGCA---------- (((...((((((((((........((((((((((.....((((.....)))))))))))))).......))))))..----))))....)))..---------- ( -28.56, z-score = -3.04, R) >droEre2.scaffold_4929 4131536 90 - 26641161 CGUAAAUUUCGAGUGGAAUUUAUAGCCGCUUUAUAUUUGGCAUCAAAUAUGCAUGAGGCGGCGUUUAAGCCACUCCA----AAAAAAAAGCGCA---------- ......(((.((((((........((((((((((.....((((.....)))))))))))))).......)))))).)----))...........---------- ( -27.26, z-score = -2.74, R) >droAna3.scaffold_12943 640903 100 + 5039921 CGUAAAUUUCGAGUGGAAUUUAUAGCCGCUUUAUAUUUGGCAUCAAAUAUGCAUGAGGCGGCGUUUAAGCCACUCCA----CAUAGGAGCAGAGACAAAAAAAA .(((((((((....))))))))).((((((((((.....((((.....))))))))))))))((((..((...(((.----....)))))..))))........ ( -28.70, z-score = -3.12, R) >dp4.chr4_group3 9816216 88 + 11692001 CGUAAAUUUCAAGUAGAAUUUAUAGCCGCUUUAUAUUUGGCAUCAAAUAUGCAUGAGUCGGCGUUUAAGCCACUCCAA---AAAAAGACCC------------- .((((((((......))))))))....((...(((((((....)))))))))..((((.(((......)))))))...---..........------------- ( -17.10, z-score = -1.54, R) >droPer1.super_1 6934952 88 + 10282868 CGUAAAUUUCAAGUAGAAUUUAUAGCCGCUUUAUAUUUGGCAUCAAAUAUGCAUGAGUCGGCGUUUAAGCCACUCCAA---AAAAAGACCC------------- .((((((((......))))))))....((...(((((((....)))))))))..((((.(((......)))))))...---..........------------- ( -17.10, z-score = -1.54, R) >droWil1.scaffold_180703 2067139 104 - 3946847 CGUAAAUUUCGAGUAGAAUUUAUAGCCGCUUUAUAUUUGGCAUCAAAUAUGCAUGAGACGGCAUUUAAGCCAGUUCCAGCCAACAAGAAUGGUGGUCAGGGAAG .((((((((......)))))))).(((((((((((((((....))))))))...))).))))...........((((.((((.((....)).))))...)))). ( -25.30, z-score = -1.53, R) >droMoj3.scaffold_6500 2161027 98 - 32352404 CGUAAAUUUCGAGUAGAAUUUAUGGCCGCUUUAUAUUUGGCAUCAAAUAUGCAUGAGCCAGCGUUUAAGCCAGUUUCCAAGUUCCAAUUUCCCGAGCU------ (((((((((......)))))))))...((((.....(((((.(((........)))))))).....)))).........(((((.........)))))------ ( -18.40, z-score = -0.34, R) >droGri2.scaffold_15252 12654107 92 - 17193109 CGUAAAUUUCGAGUAGAAUUUAUGGCCGCUUUAUAUUUGGCAUCAAAUAUGCAUGAGGCAGCGUUUAAGCCUCAGACAGUUUCCCAAUUUGA------------ (((((((((......)))))))))((((.........)))).((((((.((..((((((.........))))))..))........))))))------------ ( -20.30, z-score = -0.81, R) >droVir3.scaffold_12963 11041875 90 + 20206255 CGUAAAUUUCGAGUUGAAUUUAUGGCCGCUUUAUAUUUGGCAUCAAAUAUGCAUGAGCCAGCGUUUAAGCCAGUUCC--AGUACCAAUUUCA------------ (((((((((......)))))))))...((((.....(((((.(((........)))))))).....)))).......--.............------------ ( -16.30, z-score = -0.14, R) >consensus CGUAAAUUUCGAGUGGAAUUUAUAGCCGCUUUAUAUUUGGCAUCAAAUAUGCAUGAGGCGGCGUUUAAGCCACUCCA____AAAAAAAAUCGC___________ .((((((((......)))))))).((.((((((((((((....))))))))...))).).)).......................................... (-11.48 = -11.48 + -0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:15:17 2011