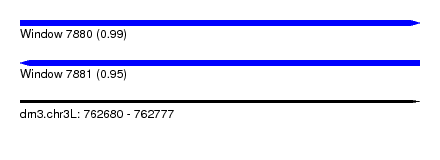
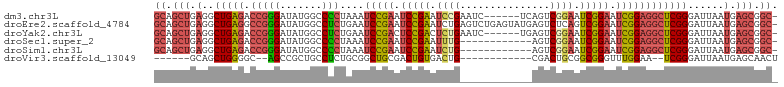
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 762,680 – 762,777 |
| Length | 97 |
| Max. P | 0.994502 |

| Location | 762,680 – 762,777 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 75.68 |
| Shannon entropy | 0.43632 |
| G+C content | 0.57891 |
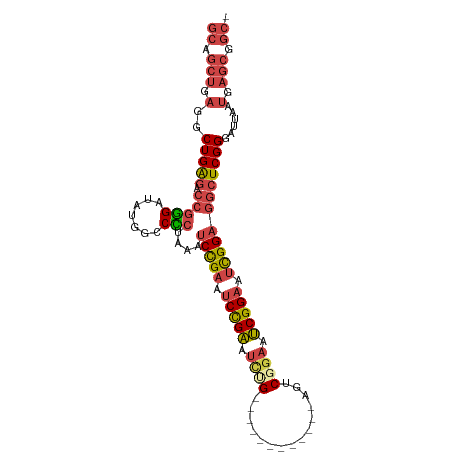
| Mean single sequence MFE | -34.42 |
| Consensus MFE | -21.43 |
| Energy contribution | -22.33 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.71 |
| SVM RNA-class probability | 0.994502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
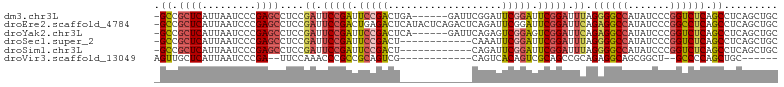
>dm3.chr3L 762680 97 + 24543557 -GCCGCUCAUUAAUCCCGAGCCUCCGAUUCCGAUUCCGACUGA------GAUUCGGAUUCGGAUUCGGAUUUAGGGGCCAUAUCCCGGUCUCAGCCUCAGCUGC -(((((((.........)))).(((((.(((((.(((((....------...))))).))))).)))))....((((.....)))))))..((((....)))). ( -38.10, z-score = -2.32, R) >droEre2.scaffold_4784 775510 103 + 25762168 -GCCGCUCAUUAAUCCCGAGCCUCCGAUUCCGACUGAGACUCAUACUCAGACUCAGAUUCGGAUUCGGAUUCAGAGGCCAUAUCCCGGCCUCAGCCUCAGCUGC -...((((.........)))).(((((.((((((((((.((.......)).)))))..))))).)))))....((((((.......))))))(((....))).. ( -38.40, z-score = -3.62, R) >droYak2.chr3L 756953 97 + 24197627 -GCCGCUCAUUAAUCCCGAGCCUCCGAUUCCGAUUCCGACUCA------GAUUCAGAGUCGGAGUCGGAUUCAGAGGCCAUAUCCCGGUCUCAGCCUCAGCUGC -((.(((..........(.(((((.((.((((((((((((((.------......)))))))))))))).)).)))))).......(((....)))..))).)) ( -44.50, z-score = -5.52, R) >droSec1.super_2 782303 91 + 7591821 -GCCGCUCAUUAAUCCCGAGCCUCCGAUUCCGAUUCCGACU------------CAAAUUCGGAUUCGGAUUUAGGGGCCAUAUCCCGGUCUCAGCCUCAGCUGC -((.(((..........(.(((((..(.(((((.(((((..------------.....))))).))))).)..)))))).......(((....)))..))).)) ( -31.30, z-score = -2.23, R) >droSim1.chr3L 413735 91 + 22553184 -GCCGCUCAUUAAUCCCGAGCCUCCGAUUCCGAUUCCGACU------------CAGAUUCGGAUUCGGAUUUAGGGGCCAUAUCCCGGUCUCAGCCUCAGCUGC -((.(((..........(.(((((..(.(((((.(((((..------------.....))))).))))).)..)))))).......(((....)))..))).)) ( -31.30, z-score = -1.72, R) >droVir3.scaffold_13049 16371007 82 - 25233164 AGUUGCUCAUUAAUCCCGA--UUCCAAACCCGCCGCAGUCG------------CAGUCACAGUCGCAGCCGCAGAGGCAGCGGCU--GCCCCAGCUGC------ .((.((.............--..........)).))....(------------((((..(((((((.(((.....))).))))))--).....)))))------ ( -22.90, z-score = -0.28, R) >consensus _GCCGCUCAUUAAUCCCGAGCCUCCGAUUCCGAUUCCGACU____________CAGAUUCGGAUUCGGAUUCAGAGGCCAUAUCCCGGUCUCAGCCUCAGCUGC .((.(((............(((((.((.(((((.(((((...................))))).))))).)).)))))........(((....)))..))).)) (-21.43 = -22.33 + 0.89)
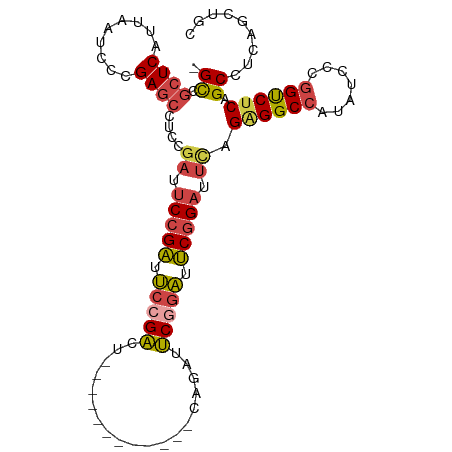
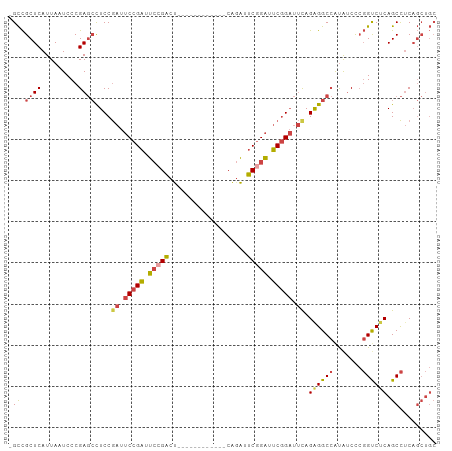
| Location | 762,680 – 762,777 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 75.68 |
| Shannon entropy | 0.43632 |
| G+C content | 0.57891 |
| Mean single sequence MFE | -38.25 |
| Consensus MFE | -22.67 |
| Energy contribution | -24.09 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.948233 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 762680 97 - 24543557 GCAGCUGAGGCUGAGACCGGGAUAUGGCCCCUAAAUCCGAAUCCGAAUCCGAAUC------UCAGUCGGAAUCGGAAUCGGAGGCUCGGGAUUAAUGAGCGGC- ((.(((.(..(((((.(((((.......)))....(((((.(((((.(((((...------....))))).))))).))))))))))))......).))).))- ( -40.00, z-score = -2.12, R) >droEre2.scaffold_4784 775510 103 - 25762168 GCAGCUGAGGCUGAGGCCGGGAUAUGGCCUCUGAAUCCGAAUCCGAAUCUGAGUCUGAGUAUGAGUCUCAGUCGGAAUCGGAGGCUCGGGAUUAAUGAGCGGC- ....(((((.(.(((((((.....)))))))....(((((.(((((..(((((.((.......)).)))))))))).)))))).)))))..............- ( -43.70, z-score = -2.52, R) >droYak2.chr3L 756953 97 - 24197627 GCAGCUGAGGCUGAGACCGGGAUAUGGCCUCUGAAUCCGACUCCGACUCUGAAUC------UGAGUCGGAAUCGGAAUCGGAGGCUCGGGAUUAAUGAGCGGC- ((.(((.(...(((..(((......(((((((((.(((((.((((((((......------.)))))))).))))).))))))))))))..))).).))).))- ( -49.70, z-score = -5.16, R) >droSec1.super_2 782303 91 - 7591821 GCAGCUGAGGCUGAGACCGGGAUAUGGCCCCUAAAUCCGAAUCCGAAUUUG------------AGUCGGAAUCGGAAUCGGAGGCUCGGGAUUAAUGAGCGGC- ((.(((.(..(((((.(((((.......)))....(((((.(((((.((((------------...)))).))))).))))))))))))......).))).))- ( -32.60, z-score = -1.21, R) >droSim1.chr3L 413735 91 - 22553184 GCAGCUGAGGCUGAGACCGGGAUAUGGCCCCUAAAUCCGAAUCCGAAUCUG------------AGUCGGAAUCGGAAUCGGAGGCUCGGGAUUAAUGAGCGGC- ((.(((.(..(((((.(((((.......)))....(((((.(((((.((((------------...)))).))))).))))))))))))......).))).))- ( -35.30, z-score = -1.71, R) >droVir3.scaffold_13049 16371007 82 + 25233164 ------GCAGCUGGGGC--AGCCGCUGCCUCUGCGGCUGCGACUGUGACUG------------CGACUGCGGCGGGUUUGGAA--UCGGGAUUAAUGAGCAACU ------(((((.(..((--((((((.......))))))))..)...).)))------------)...(((....(((((.(..--.).))))).....)))... ( -28.20, z-score = 0.69, R) >consensus GCAGCUGAGGCUGAGACCGGGAUAUGGCCCCUAAAUCCGAAUCCGAAUCUG____________AGUCGGAAUCGGAAUCGGAGGCUCGGGAUUAAUGAGCGGC_ ((.(((.(..(((((.(((((.......)))....(((((.(((((.((((...............)))).))))).))))))))))))......).))).)). (-22.67 = -24.09 + 1.42)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:54:07 2011