
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 755,195 – 755,297 |
| Length | 102 |
| Max. P | 0.772133 |

| Location | 755,195 – 755,297 |
|---|---|
| Length | 102 |
| Sequences | 10 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 75.72 |
| Shannon entropy | 0.46508 |
| G+C content | 0.47338 |
| Mean single sequence MFE | -25.54 |
| Consensus MFE | -10.22 |
| Energy contribution | -10.22 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.772133 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
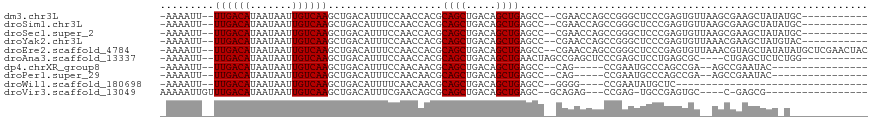
>dm3.chr3L 755195 102 + 24543557 -AAAAUU--UUGACAUAAUAAUUGUCAAGCUGACAUUUCCAACCACGCAGCUGACAGCUGAGCC--CGAACCAGCCGGGCUCCCGAGUGUUAAGCGAAGCUAUAUGC----------- -......--((((((.......))))))((((((((((..........(((.....)))(((((--((.......)))))))..))))))).)))............----------- ( -29.60, z-score = -2.77, R) >droSim1.chr3L 405637 102 + 22553184 -AAAAUU--UUGACAUAAUAAUUGUCAAGCUGACAUUUCCAACCACGCAGCUGACAGCUGAGCC--CGAACCAGCCGGGCUCCCGAGUGUUAAGCGAAGCUAUAUGC----------- -......--((((((.......))))))((((((((((..........(((.....)))(((((--((.......)))))))..))))))).)))............----------- ( -29.60, z-score = -2.77, R) >droSec1.super_2 774989 102 + 7591821 -AAAAUU--UUGACAUAAUAAUUGUCAAGCUGACAUUUCCAACCACGCAGCUGACAGCUGAGCC--CGAACCAGCCGGGCUCCCGAGUGUUAAGCGAAGCUAUAUGC----------- -......--((((((.......))))))((((((((((..........(((.....)))(((((--((.......)))))))..))))))).)))............----------- ( -29.60, z-score = -2.77, R) >droYak2.chr3L 749052 102 + 24197627 -AAAAUU--UUGACAUAAUAAUUGUCAAGCUGACAUUUCCAACCACGCAGCUGACAGCUGAGCC--CGAACCAGCCGGGCUCCCGAGUGUUAAACGAAGCUAUGUAC----------- -.....(--(((((((.....((((((.((((.(............)))))))))))..(((((--((.......)))))))....)))))))).............----------- ( -29.20, z-score = -3.25, R) >droEre2.scaffold_4784 767740 113 + 25762168 -AAAAUU--UUGACAUAAUAAUUGUCAAGCUGACAUUUCCAACCACGCAGCUGACAGCUGAGCC--CGAACCAGCCGGGCUCCCGAGUGUUAAACGUAGCUAUAUAUGCUCGAACUAC -.....(--(((((((.....((((((.((((.(............)))))))))))..(((((--((.......)))))))....))))))))((.(((.......)))))...... ( -33.40, z-score = -3.88, R) >droAna3.scaffold_13337 11705231 100 + 23293914 -AAAAUU--UUGACAUAAUAAUUGUCAAGCUGACAUUUCCAACCACGCAGCUGACAGCUGAACUAGCCGAGCUCCCGAGCUCCUGAGCGC----CUGAGCUCUCUGG----------- -.....(--((((((.......)))))))..................((((.....))))..((((..((((((....(((....)))..----..)))))).))))----------- ( -26.50, z-score = -1.72, R) >dp4.chrXR_group8 4729735 90 - 9212921 -AAAAUU--UUGACAUAAUAAUUGUCAAGCUGACAUUUCCAACAACGCAGCUGACAGCUGAGCC--CAG-----CCGAAUGCCCAGCCGA--AGCCGAAUAC---------------- -......--((((((.......))))))((((.(((((.........((((.....))))....--...-----..)))))..))))...--..........---------------- ( -18.67, z-score = -2.13, R) >droPer1.super_29 380813 90 - 1099123 -AAAAUU--UUGACAUAAUAAUUGUCAAGCUGACAUUUCCAACAACGCAGCUGACAGCUGAGCC--CAG-----CCGAAUGCCCAGCCGA--AGCCGAAUAC---------------- -......--((((((.......))))))((((.(((((.........((((.....))))....--...-----..)))))..))))...--..........---------------- ( -18.67, z-score = -2.13, R) >droWil1.scaffold_180698 8959241 77 + 11422946 -AAAAUU--UUGACAUAAUAAUUGUCAAGCUGACAUUUUCAACAACGCAGCUGACAGCUGAGCC--GGGG----CCGAAUAUGCUC-------------------------------- -.....(--((((((.......)))))))....(((.(((.......((((.....)))).((.--...)----).))).)))...-------------------------------- ( -15.70, z-score = -0.38, R) >droVir3.scaffold_13049 16362355 90 - 25233164 AAAAAUUGUUUGACAUAAUAAUUGUCAAGCUGACAUUUCGAACAGCGCAGCUGACAGCUGAGC--GCAGAG---CCGAG-UGCCGAGUGC----C-GAGCG----------------- .......((((((((.......))))))))..............(((((((.....)))..))--))...(---(((.(-..(...)..)----)-).)).----------------- ( -24.50, z-score = -0.34, R) >consensus _AAAAUU__UUGACAUAAUAAUUGUCAAGCUGACAUUUCCAACCACGCAGCUGACAGCUGAGCC__CGAACCAGCCGAGCUCCCGAGUGU__AGCGAAGCUAU_U_____________ .........((((((.......))))))...................((((.....)))).......................................................... (-10.22 = -10.22 + -0.00)

| Location | 755,195 – 755,297 |
|---|---|
| Length | 102 |
| Sequences | 10 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 75.72 |
| Shannon entropy | 0.46508 |
| G+C content | 0.47338 |
| Mean single sequence MFE | -29.96 |
| Consensus MFE | -13.98 |
| Energy contribution | -14.00 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
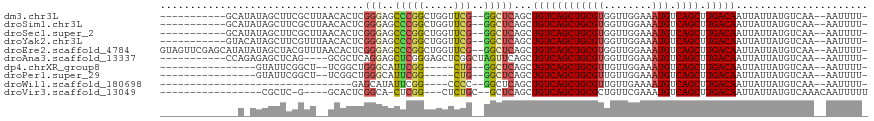
>dm3.chr3L 755195 102 - 24543557 -----------GCAUAUAGCUUCGCUUAACACUCGGGAGCCCGGCUGGUUCG--GGCUCAGCUGUCAGCUGCGUGGUUGGAAAUGUCAGCUUGACAAUUAUUAUGUCAA--AAUUUU- -----------(((((.(((((((.........)))((((((((.....)))--)))))))))(((((((((((........))).)))).))))......)))))...--......- ( -31.30, z-score = -1.20, R) >droSim1.chr3L 405637 102 - 22553184 -----------GCAUAUAGCUUCGCUUAACACUCGGGAGCCCGGCUGGUUCG--GGCUCAGCUGUCAGCUGCGUGGUUGGAAAUGUCAGCUUGACAAUUAUUAUGUCAA--AAUUUU- -----------(((((.(((((((.........)))((((((((.....)))--)))))))))(((((((((((........))).)))).))))......)))))...--......- ( -31.30, z-score = -1.20, R) >droSec1.super_2 774989 102 - 7591821 -----------GCAUAUAGCUUCGCUUAACACUCGGGAGCCCGGCUGGUUCG--GGCUCAGCUGUCAGCUGCGUGGUUGGAAAUGUCAGCUUGACAAUUAUUAUGUCAA--AAUUUU- -----------(((((.(((((((.........)))((((((((.....)))--)))))))))(((((((((((........))).)))).))))......)))))...--......- ( -31.30, z-score = -1.20, R) >droYak2.chr3L 749052 102 - 24197627 -----------GUACAUAGCUUCGUUUAACACUCGGGAGCCCGGCUGGUUCG--GGCUCAGCUGUCAGCUGCGUGGUUGGAAAUGUCAGCUUGACAAUUAUUAUGUCAA--AAUUUU- -----------(.(((((((((((.........)))((((((((.....)))--))))))))((((((((((((........))).)))).))))).....))))))..--......- ( -32.60, z-score = -2.07, R) >droEre2.scaffold_4784 767740 113 - 25762168 GUAGUUCGAGCAUAUAUAGCUACGUUUAACACUCGGGAGCCCGGCUGGUUCG--GGCUCAGCUGUCAGCUGCGUGGUUGGAAAUGUCAGCUUGACAAUUAUUAUGUCAA--AAUUUU- ...(((.((.(((...(((((((((...........((((((((.....)))--)))))(((.....))))))))))))...))))))))((((((.......))))))--......- ( -35.30, z-score = -1.81, R) >droAna3.scaffold_13337 11705231 100 - 23293914 -----------CCAGAGAGCUCAG----GCGCUCAGGAGCUCGGGAGCUCGGCUAGUUCAGCUGUCAGCUGCGUGGUUGGAAAUGUCAGCUUGACAAUUAUUAUGUCAA--AAUUUU- -----------.....((((((..----(.(((....))).)..))))))(((((((.....((((((((((((........))).)))).)))))...)))).)))..--......- ( -31.90, z-score = -0.65, R) >dp4.chrXR_group8 4729735 90 + 9212921 ----------------GUAUUCGGCU--UCGGCUGGGCAUUCGG-----CUG--GGCUCAGCUGUCAGCUGCGUUGUUGGAAAUGUCAGCUUGACAAUUAUUAUGUCAA--AAUUUU- ----------------.....((((.--...))))(((((..((-----(((--....)))))((((((((((((......)))).)))).)))).......)))))..--......- ( -31.00, z-score = -2.98, R) >droPer1.super_29 380813 90 + 1099123 ----------------GUAUUCGGCU--UCGGCUGGGCAUUCGG-----CUG--GGCUCAGCUGUCAGCUGCGUUGUUGGAAAUGUCAGCUUGACAAUUAUUAUGUCAA--AAUUUU- ----------------.....((((.--...))))(((((..((-----(((--....)))))((((((((((((......)))).)))).)))).......)))))..--......- ( -31.00, z-score = -2.98, R) >droWil1.scaffold_180698 8959241 77 - 11422946 --------------------------------GAGCAUAUUCGG----CCCC--GGCUCAGCUGUCAGCUGCGUUGUUGAAAAUGUCAGCUUGACAAUUAUUAUGUCAA--AAUUUU- --------------------------------((.((((...((----(...--.)))....(((((((((((((......)))).)))).))))).....))))))..--......- ( -19.80, z-score = -1.27, R) >droVir3.scaffold_13049 16362355 90 + 25233164 -----------------CGCUC-G----GCACUCGGCA-CUCGG---CUCUGC--GCUCAGCUGUCAGCUGCGCUGUUCGAAAUGUCAGCUUGACAAUUAUUAUGUCAAACAAUUUUU -----------------.(((.-(----(((.((((((-.....---...)))--((.((((.....)))).))....)))..)))))))((((((.......))))))......... ( -24.10, z-score = -0.49, R) >consensus _____________A_AUAGCUCCGCU__ACACUCGGGAGCCCGGCUGGUCCG__GGCUCAGCUGUCAGCUGCGUGGUUGGAAAUGUCAGCUUGACAAUUAUUAUGUCAA__AAUUUU_ ................................((((....))))..................((((((((((((........))).)))).)))))...................... (-13.98 = -14.00 + 0.02)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:54:04 2011