| Sequence ID | dm3.chr3L |
|---|---|
| Location | 707,315 – 707,424 |
| Length | 109 |
| Max. P | 0.640324 |

| Location | 707,315 – 707,424 |
|---|---|
| Length | 109 |
| Sequences | 11 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 69.22 |
| Shannon entropy | 0.64878 |
| G+C content | 0.54386 |
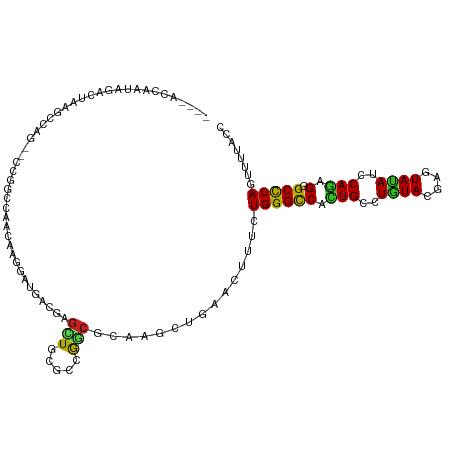
| Mean single sequence MFE | -34.46 |
| Consensus MFE | -14.20 |
| Energy contribution | -13.43 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.56 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.640324 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
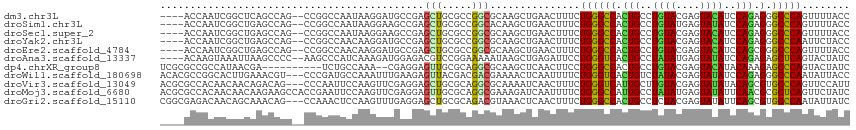
>dm3.chr3L 707315 109 - 24543557 ----ACCAAUCGGCUCAGCCAG--CCGGCCAAUAAGGAUGCCGAGCUGCGCCGGCGCAAGCUGAACUUUCUGGGCCACUGCCUGUACGAGUACAUCCAGAGGGCCCAGUUUUACC ----......((((.(((((.(--(...((.....))..)).).)))).))))((....))........(((((((.(((..((((....))))..)))..)))))))....... ( -44.00, z-score = -2.05, R) >droSim1.chr3L 356699 109 - 22553184 ----ACCAAUCGGCUGAGCCAG--CCGGCCAAUAAGGAAGCCGAGCUGCGCCGGCACAAGCUGAACUUUCUGGGCCACUGCCUGUAUGAGUAUAUCCAGAGGGCCCAGUUUUACC ----........((((.(((((--(((((..........)))).)))).))))))..............(((((((.(((..((((....))))..)))..)))))))....... ( -40.10, z-score = -1.35, R) >droSec1.super_2 727238 109 - 7591821 ----ACCAAUCGGCUGAGCCAG--CCGGCCAAUAAGGAAGCCGAGCUGCGCCGGCGCAAGCUGAACUUUCUGGGCCACUGCCUGUACGAGUACAUCCAGAGGGCCCAGUUUUACC ----.....((((((..((..(--(((((((....((...))....)).)))))))).)))))).....(((((((.(((..((((....))))..)))..)))))))....... ( -45.10, z-score = -2.31, R) >droYak2.chr3L 699117 109 - 24197627 ----ACCAAUCGGCUGAGCCAG--CCGGCCAACAAGGAUGCCGAGCUGCGCCGGCGCAAGCUGAACUUUCUGGGCCACUGCCUGUACGAGUACAUCCAGAGGGCCCAAUUCUACC ----.....((((((..((..(--(((((((.(..((...))..).)).)))))))).))))))......((((((.(((..((((....))))..)))..))))))........ ( -43.20, z-score = -1.88, R) >droEre2.scaffold_4784 717232 109 - 25762168 ----ACCAAUCGGCUGAGCCAG--CCGGCCAACAAGGAUGCCGAGCUGCGCCGGCGCAAGCUGAACUUUCUGGGCCACUGCCUGUACGAGUACAUCCAGCGGGCCCAGUUUUACC ----.....((((((..((..(--(((((((.(..((...))..).)).)))))))).)))))).....(((((((.(((..((((....))))..)))..)))))))....... ( -45.10, z-score = -1.84, R) >droAna3.scaffold_13337 11653425 109 - 23293914 ----ACAAGUAAAUUAAGCCCC--AAGCCCAUCAAAGAUGGAGACGUCCGGAAAAAUAAGCUGAGAUUCCUGGGUCACUGCCUAUAUGAGUAUAUCCAGAGAGCUCAGUACUAUC ----...(((............--.......((...((((....))))..)).......((((((.(((((((((...((((.....).))).)))))).))))))))))))... ( -26.00, z-score = -1.54, R) >dp4.chrXR_group8 4679818 103 + 9212921 UCGCGCCGCCAUAACGA----------UCUGCCAAA--CGAGGAGUUGCGCAGGCGCAAGCUCAACUUCCUGGGCCACUGCCUGUACGAGUACAUACAAAGAGCCCAGUACUAUC ..(((((((..((((..----------((.......--.))...)))).)).)))))............(((((((..((..((((....))))..))..).))))))....... ( -29.70, z-score = -0.52, R) >droWil1.scaffold_180698 8899146 112 - 11422946 ACACGCCGGCACUUGAAACGU---CCCGAUGCCAAAUUUGAAGAGUUACGACGACGAAAACUCAAUUUUCUGGGUCACUGUCUAUACGAGUAUAUCCAGAGGGCCCAAUAUUACC .......((((.(((......---..))))))).........(((((.((....))..))))).......((((((.(((..((((....))))..)))..))))))........ ( -26.10, z-score = -0.90, R) >droVir3.scaffold_13049 16304998 112 + 25233164 ACGCGCCACAACAACAGACAG---CCCAAUUCCAAGUUCGAGGAGCUGCGCAGGCGCAAAAUCAACUUUCUGGGUCAUUGCUUGUACGAGUAUAUACAGCGUGCCCAGUUCCAUU ..(((((....(....)...(---(.((.((((........)))).)).)).)))))............(((((.(((.(((.(((........))))))))))))))....... ( -30.70, z-score = -1.13, R) >droMoj3.scaffold_6680 6417440 115 - 24764193 ACGCGCCACAACAACAAGAAGCCACCGAAUUCCAAGUUCGAGGAGUUGCGCAGGCGAAAGAUCAAUUUUCUGGGCCAUUGCCUAUAUGAGUAUAUUCAACGCGCUCAGUUCUAUC ..((((..............(((..((((((((........)))))).))..)))....((...((((..(((((....)))))...))))....))...))))........... ( -25.80, z-score = -0.03, R) >droGri2.scaffold_15110 19874248 112 + 24565398 CGGCGAGACAACAGCAAACAG---CCAAACUCCAAGUUUGAGGAGCUGCGCAGACGUAAACUCAACUUUCUGGGCCACUGCCUCUACGAGUAUAUUCAGCGUGCCCAAUAUUAUC .((((.............(((---((((((.....)))))....))))(((.((.(((.((((..(.....)(((....))).....)))).))))).))))))).......... ( -23.30, z-score = 0.82, R) >consensus ____ACCAAUAGACUAAGCCAG__CCGGCCAACAAGGAUGACGAGCUGCGCCGGCGCAAGCUGAACUUUCUGGGCCACUGCCUGUACGAGUAUAUCCAGAGGGCCCAGUUUUACC ............................................(((.....)))...............((((((.(((..((((....))))..))).).)))))........ (-14.20 = -13.43 + -0.77)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:53:59 2011