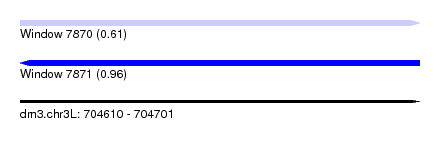
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 704,610 – 704,701 |
| Length | 91 |
| Max. P | 0.964021 |

| Location | 704,610 – 704,701 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 81.39 |
| Shannon entropy | 0.35727 |
| G+C content | 0.48132 |
| Mean single sequence MFE | -25.56 |
| Consensus MFE | -14.73 |
| Energy contribution | -16.71 |
| Covariance contribution | 1.98 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.606873 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
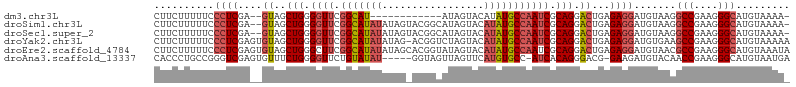
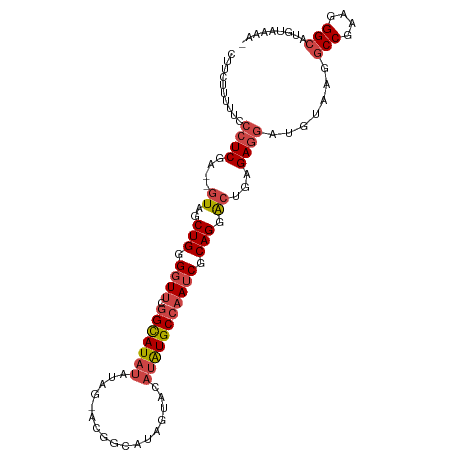
>dm3.chr3L 704610 91 + 24543557 -UUUUACAUGCCCUUCGGCCUUACAUCCUCUCAGUCCUGCGAUUGGCAUAUGUACUAU------------AUGCCGAACCCCAGCUAC--UCGAGGGAAAAAGAAG -........(((....)))(((...(((.((((((.(((...((((((((((...)))------------)))))))....)))..))--).))))))..)))... ( -23.40, z-score = -2.06, R) >droSim1.chr3L 353998 103 + 22553184 -UUUUACAUGCCCUUCGGCCUUACAUCCUCUCAGUCCUGCGAUUGGCAUAUGUACUAUGCCGUACUAUAUAUGCCGAACCCCAGCUAC--UCGAGGGAAAAAGAAG -........(((....)))(((...(((.((((((.(((...((((((((((((.(((...))).))))))))))))....)))..))--).))))))..)))... ( -27.10, z-score = -2.36, R) >droSec1.super_2 724535 103 + 7591821 -UUUUACAUGCCCUUCGGCCUUACAUCCUCUCAGUCCUGCGAUUGGCAUAUGUACUAUGCCGUACUAUAUAUGCCGAACCCCAGCUAC--UCGAGGGAAAAAGAAG -........(((....)))(((...(((.((((((.(((...((((((((((((.(((...))).))))))))))))....)))..))--).))))))..)))... ( -27.10, z-score = -2.36, R) >droYak2.chr3L 696357 105 + 24197627 UUUUUACAUGCCCUUCGGCUUCACAUCCUCUCAGUCCUGCGAUUGGCAUAUGUACUAGACCGU-CUAUAUAUGCCGAACCCCAGCUACACUCGAGGGAAAAAGAAG .........(((....)))......(((.((((((...((..((((((((((((.........-.))))))))))))......))...))).))))))........ ( -26.00, z-score = -1.92, R) >droEre2.scaffold_4784 714508 106 + 25762168 UAUUUACAUGCCCUUCGGCGUUACAUCCUCUCAGUCCUGCGAUUGGCAUAUGUACUAUACCGUGCUAUAUAUGCCGAAGCCCAGCUACACUCGAGGGAAAAAGAAG .......(((((....))))).......(((...((((.((((((((((((((((........).))))))))))))(((...)))....))).))))...))).. ( -28.80, z-score = -1.91, R) >droAna3.scaffold_13337 11649531 99 + 23293914 UCAUUACAUGCCCUUCGGUUGUACAUCUUC-CGUCCCUGUGAU-GGCACAUGAACUAACUACC-----AUAUACAGAACCCCAGAAACACUCGACCCGGCAGGGUG .........((((((((((((....(((..-.((..(((((((-((...............))-----)).))))).))...)))......))).)))).))))). ( -20.96, z-score = -0.04, R) >consensus _UUUUACAUGCCCUUCGGCCUUACAUCCUCUCAGUCCUGCGAUUGGCAUAUGUACUAUACCGU_CUAUAUAUGCCGAACCCCAGCUAC__UCGAGGGAAAAAGAAG .........(((....))).........(((...((((.(((((((((((((((...........)))))))))))).............))).))))...))).. (-14.73 = -16.71 + 1.98)

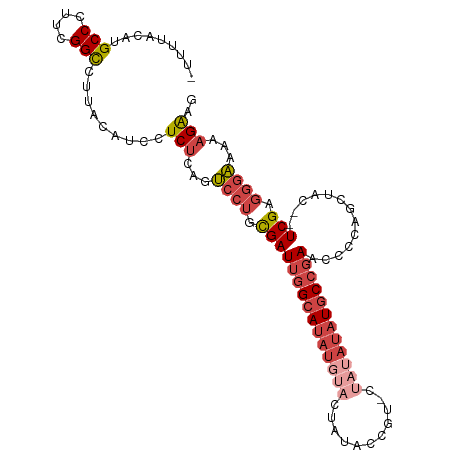
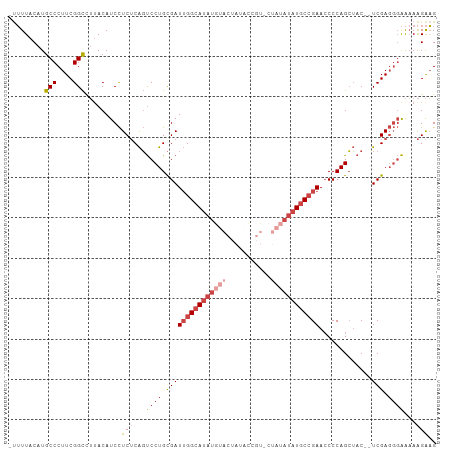
| Location | 704,610 – 704,701 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 81.39 |
| Shannon entropy | 0.35727 |
| G+C content | 0.48132 |
| Mean single sequence MFE | -34.97 |
| Consensus MFE | -21.76 |
| Energy contribution | -22.85 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.964021 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 704610 91 - 24543557 CUUCUUUUUCCCUCGA--GUAGCUGGGGUUCGGCAU------------AUAGUACAUAUGCCAAUCGCAGGACUGAGAGGAUGUAAGGCCGAAGGGCAUGUAAAA- ...(((..((((((.(--((..(((.((((.(((((------------((.....))))))))))).))).)))))).)))...)))(((....)))........- ( -34.70, z-score = -3.42, R) >droSim1.chr3L 353998 103 - 22553184 CUUCUUUUUCCCUCGA--GUAGCUGGGGUUCGGCAUAUAUAGUACGGCAUAGUACAUAUGCCAAUCGCAGGACUGAGAGGAUGUAAGGCCGAAGGGCAUGUAAAA- ...(((..((((((.(--((..(((.((((.(((((((...((((......))))))))))))))).))).)))))).)))...)))(((....)))........- ( -38.80, z-score = -3.67, R) >droSec1.super_2 724535 103 - 7591821 CUUCUUUUUCCCUCGA--GUAGCUGGGGUUCGGCAUAUAUAGUACGGCAUAGUACAUAUGCCAAUCGCAGGACUGAGAGGAUGUAAGGCCGAAGGGCAUGUAAAA- ...(((..((((((.(--((..(((.((((.(((((((...((((......))))))))))))))).))).)))))).)))...)))(((....)))........- ( -38.80, z-score = -3.67, R) >droYak2.chr3L 696357 105 - 24197627 CUUCUUUUUCCCUCGAGUGUAGCUGGGGUUCGGCAUAUAUAG-ACGGUCUAGUACAUAUGCCAAUCGCAGGACUGAGAGGAUGUGAAGCCGAAGGGCAUGUAAAAA .(((....((((((.(((....(((.((((.(((((((.(((-.....)))....))))))))))).))).)))))).)))...)))(((....)))......... ( -33.70, z-score = -1.77, R) >droEre2.scaffold_4784 714508 106 - 25762168 CUUCUUUUUCCCUCGAGUGUAGCUGGGCUUCGGCAUAUAUAGCACGGUAUAGUACAUAUGCCAAUCGCAGGACUGAGAGGAUGUAACGCCGAAGGGCAUGUAAAUA ((((((..(((..(((..((.((((.((....)).....))))))((((((.....))))))..)))..)))..)))))).......(((....)))......... ( -36.10, z-score = -2.43, R) >droAna3.scaffold_13337 11649531 99 - 23293914 CACCCUGCCGGGUCGAGUGUUUCUGGGGUUCUGUAUAU-----GGUAGUUAGUUCAUGUGCC-AUCACAGGGACG-GAAGAUGUACAACCGAAGGGCAUGUAAUGA ..((((..((((((...(((..(((.(((...((((((-----((........)))))))).-))).)))..)))-...)))......))).)))).......... ( -27.70, z-score = 0.44, R) >consensus CUUCUUUUUCCCUCGA__GUAGCUGGGGUUCGGCAUAUAUAG_ACGGCAUAGUACAUAUGCCAAUCGCAGGACUGAGAGGAUGUAAGGCCGAAGGGCAUGUAAAA_ ..........((((....((..(((.((((.(((((((.................))))))))))).))).))...)))).......(((....)))......... (-21.76 = -22.85 + 1.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:53:58 2011