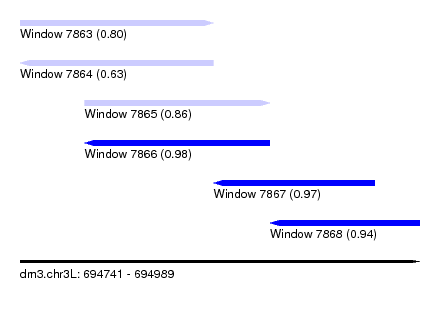
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 694,741 – 694,989 |
| Length | 248 |
| Max. P | 0.980887 |

| Location | 694,741 – 694,861 |
|---|---|
| Length | 120 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.78 |
| Shannon entropy | 0.27199 |
| G+C content | 0.58333 |
| Mean single sequence MFE | -49.98 |
| Consensus MFE | -28.14 |
| Energy contribution | -28.60 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.799192 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
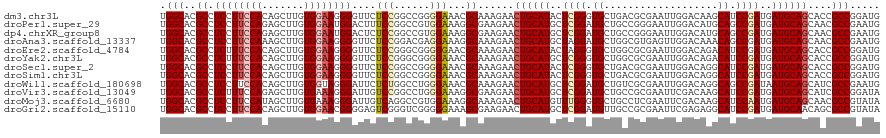
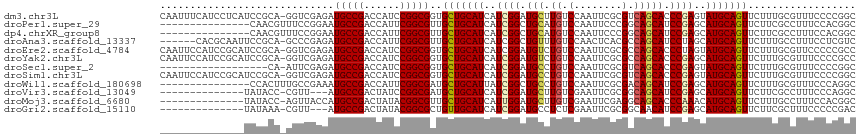
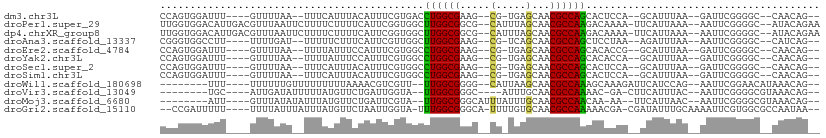
>dm3.chr3L 694741 120 + 24543557 UGGCACGCCUCCUUCCACAGCUUGUGGAAGGGGUUCUCCGGCCGGGGAAACGCAAAGAACUGCAUACUCGGGUGCUGACGCGAAUUGGACAAGCAUCCGAUGAUGCAGCACCGCCGGAUG ......(((.((((((((.....)))))))))))..((((((.(((....)........((((((..(((((((((....(......)...)))))))))..))))))..)))))))).. ( -56.20, z-score = -4.13, R) >droPer1.super_29 316470 120 - 1099123 UGGCACGCCUCCUUCCAGAGCUUGUGGAAUGGACUUUCCGGCCGUGGAAAGGCGAAGAACUGCAUGCUCGGAUGCUGCCGGGAAUUGGACAUGCAGCCGAUGAUGCAGCAACGCCGAAUG .(((.(((((..(((((..(((...((((......)))))))..)))))))))).....((((((..((((.((((((((.....))).)).))).))))..))))))....)))..... ( -47.30, z-score = -2.00, R) >dp4.chrXR_group8 4666411 120 - 9212921 UGGCACGCCUCCUUCCAGAGCUUGUGGAAUGGACUCUCCGGCCGUGGAAAGGCGAAGAACUGCAUGCUCGGAUGCUGCCGGGAAUUGGACAUGCAGCCGAUGAUGCAGCAACGCCGAAUG .(((.(((((..(((((..(((...(((........))))))..)))))))))).....((((((..((((.((((((((.....))).)).))).))))..))))))....)))..... ( -45.60, z-score = -1.36, R) >droAna3.scaffold_13337 11638923 120 + 23293914 UGGCACGCCUCCUUCCAAAGCUUGUGGAAGGGGUUCUCCGGACGAGGAAAGGCAAAGAACUGCAUGCUAGGAUGCUGGCGUGAGUUGGACAAACAGCCGAUGAUGCAGCAACGCCGAAUG .(((..(((((((((((.......)))))))..(((((.....))))).))))......(((((((((.(..(((..((....))..).))..)))).....))))))....)))..... ( -43.10, z-score = -1.41, R) >droEre2.scaffold_4784 704292 120 + 25762168 UGGCACGCCUCUUUCCACAGCUUGUGGAAGGGGUUCUCCGGGCGGGGGAACGCAAAGAACUGCAUACUAGGGUGCUGGCGCGAAUUGGACAGACAUCCGAUGAUGCAGCACCGCCGGAUG .(((..((.(((((((((.....)))))))))(((((((.....))))))))).................(((((((.((...((((((......))))))..))))))))))))..... ( -49.00, z-score = -2.13, R) >droYak2.chr3L 684707 120 + 24197627 UGGCACGCCUCUUUCCACAGCUUGUGGAAGGGGUUCUCCGGGCGGGGAAACGCAAAGAACUGCAUGCUCGGGUGCUGGCGCGAAUUGGACAGACAUCCGAUGAUGCAGCACCGCCGGAUG ......(((.((((((((.....)))))))))))..(((((.((((....)........((((((..((((((((((............))).)))))))..))))))..)))))))).. ( -52.50, z-score = -2.61, R) >droSec1.super_2 714739 120 + 7591821 UGGCACGCCUCCUUCCACAGCUUGUGGAAGGGGUUCUCCGGCCGGGGAAACGCAAAGAACUGCAUACUCGGGUGCUGACGCGAAUUGGACAGGCAUCCGAUGAUGCAGCACCGCCGGAUG ......(((.((((((((.....)))))))))))..((((((.(((....)........((((((..(((((((((....(......)...)))))))))..))))))..)))))))).. ( -56.20, z-score = -3.77, R) >droSim1.chr3L 344153 120 + 22553184 UGGCACGCCUCCUUCCACAGCUUGUGGAAGGGGUUCUCCGGCCGGGGAAACGCAAAGAACUGCAUACUCGGGUGCUGACGCGAAUUGGACAGGCAUCCGAUGAUGCAGCACCGCCGGAUG ......(((.((((((((.....)))))))))))..((((((.(((....)........((((((..(((((((((....(......)...)))))))))..))))))..)))))))).. ( -56.20, z-score = -3.77, R) >droWil1.scaffold_180698 8879812 120 + 11422946 UGGCACGCCUCCUUCCACAGCUUGUGGUAGGGAUUCUCUGGCCUGGGAAACGCAAAGAACUGCAUGCUCGGAUGCUGUCGCGAAUUGGACAGGCAGCCGAUAAUGCAGCAUCGCCGAAUG .(((..((.(((((((((.....)))).)))))(((((......)))))..))...((.((((((..((((.(((((((........)))).))).))))..))))))..)))))..... ( -46.10, z-score = -1.65, R) >droVir3.scaffold_13049 16290418 120 - 25233164 UGGCACGCCUCUUUCCAGAGCUUGUGAAAGGGAUUGUCCGGCCUGGGAAAGGCGAAGAACUGCAUGCUCGGAUGCUGCCGCGAAUUCGACAAGCAUCCGAUGAUGCAGCAUCGCCGGAUA .(((.(((((.(((((((.(((........((.....)))))))))))))))))..((.((((((..(((((((((....((....))...)))))))))..))))))..)))))..... ( -53.20, z-score = -3.30, R) >droMoj3.scaffold_6680 6403752 120 + 24764193 UGGCACGCCUCCUUCCAUAGCUUGUGAAAGGGAUUGUCAGGCCGUGGAAAGGCAAAGAACUGCAUGUUUGGGUGCUGCCUCGAAUUCGACAAGCAUCCAAUGAUGCAGCAACGCCGUAUA .(((..((((..(((((..(((((...((....))..)))))..)))))))))......((((((..(((((((((...(((....)))..)))))))))..))))))....)))..... ( -50.60, z-score = -3.86, R) >droGri2.scaffold_15110 19859120 120 - 24565398 UGGCACGCCUCCUUCCACAGCUUGUGGAACGGGGAGUCGGGUCGGGGGAAAGCGAAGAACUGCAUGCUCGGAUGUUGCCGCGAAUUCGAGAGGCAUCCGAUGAUGCAACAGCGCCGUAUA .(((.((.((((((((((.....)))))...))))).)).)))...((...((.......(((((..(((((((((....((....))...)))))))))..)))))...)).))..... ( -43.70, z-score = -0.26, R) >consensus UGGCACGCCUCCUUCCACAGCUUGUGGAAGGGGUUCUCCGGCCGGGGAAACGCAAAGAACUGCAUGCUCGGAUGCUGCCGCGAAUUGGACAGGCAUCCGAUGAUGCAGCACCGCCGGAUG .(((..((.((((((((.......))))))))....(((......)))...))......((((((..((((.((...................)).))))..))))))....)))..... (-28.14 = -28.60 + 0.47)
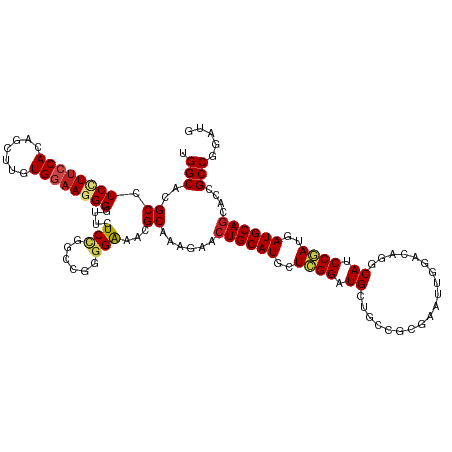
| Location | 694,741 – 694,861 |
|---|---|
| Length | 120 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.78 |
| Shannon entropy | 0.27199 |
| G+C content | 0.58333 |
| Mean single sequence MFE | -45.89 |
| Consensus MFE | -27.28 |
| Energy contribution | -26.95 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.633650 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 694741 120 - 24543557 CAUCCGGCGGUGCUGCAUCAUCGGAUGCUUGUCCAAUUCGCGUCAGCACCCGAGUAUGCAGUUCUUUGCGUUUCCCCGGCCGGAGAACCCCUUCCACAAGCUGUGGAAGGAGGCGUGCCA ..((((((((((((((......((((....)))).......).))))))).(((.((((((....)))))))))....))))))...((((((((((.....)))))))).))....... ( -49.52, z-score = -2.51, R) >droPer1.super_29 316470 120 + 1099123 CAUUCGGCGUUGCUGCAUCAUCGGCUGCAUGUCCAAUUCCCGGCAGCAUCCGAGCAUGCAGUUCUUCGCCUUUCCACGGCCGGAAAGUCCAUUCCACAAGCUCUGGAAGGAGGCGUGCCA .....((((..(((((((..((((.(((.((((........))))))).))))..)))))))....((((((((((.(((.((((......))))....))).))))..)))))))))). ( -47.80, z-score = -2.60, R) >dp4.chrXR_group8 4666411 120 + 9212921 CAUUCGGCGUUGCUGCAUCAUCGGCUGCAUGUCCAAUUCCCGGCAGCAUCCGAGCAUGCAGUUCUUCGCCUUUCCACGGCCGGAGAGUCCAUUCCACAAGCUCUGGAAGGAGGCGUGCCA .....((((..(((((((..((((.(((.((((........))))))).))))..)))))))....((((((((((.(((.((((......))))....))).))))..)))))))))). ( -47.00, z-score = -2.00, R) >droAna3.scaffold_13337 11638923 120 - 23293914 CAUUCGGCGUUGCUGCAUCAUCGGCUGUUUGUCCAACUCACGCCAGCAUCCUAGCAUGCAGUUCUUUGCCUUUCCUCGUCCGGAGAACCCCUUCCACAAGCUUUGGAAGGAGGCGUGCCA .....((((..(((((((....(((.((..(.....)..))))).((......))))))))).....((((((((......)))))...(((((((.......))))))).))).)))). ( -39.20, z-score = -1.50, R) >droEre2.scaffold_4784 704292 120 - 25762168 CAUCCGGCGGUGCUGCAUCAUCGGAUGUCUGUCCAAUUCGCGCCAGCACCCUAGUAUGCAGUUCUUUGCGUUCCCCCGCCCGGAGAACCCCUUCCACAAGCUGUGGAAAGAGGCGUGCCA .....(((((((((((......((((....)))).......).))))))).....((((.((((((((.((......)).))))))))...((((((.....))))))....))))))). ( -40.32, z-score = -1.18, R) >droYak2.chr3L 684707 120 - 24197627 CAUCCGGCGGUGCUGCAUCAUCGGAUGUCUGUCCAAUUCGCGCCAGCACCCGAGCAUGCAGUUCUUUGCGUUUCCCCGCCCGGAGAACCCCUUCCACAAGCUGUGGAAAGAGGCGUGCCA .....(((((.(((((((..((((.((.(((.((.....).).))))).))))..))))))).))..(((((((.((....))........((((((.....)))))).)))))))))). ( -41.70, z-score = -0.95, R) >droSec1.super_2 714739 120 - 7591821 CAUCCGGCGGUGCUGCAUCAUCGGAUGCCUGUCCAAUUCGCGUCAGCACCCGAGUAUGCAGUUCUUUGCGUUUCCCCGGCCGGAGAACCCCUUCCACAAGCUGUGGAAGGAGGCGUGCCA ..((((((((((((((......((((....)))).......).))))))).(((.((((((....)))))))))....))))))...((((((((((.....)))))))).))....... ( -49.52, z-score = -2.45, R) >droSim1.chr3L 344153 120 - 22553184 CAUCCGGCGGUGCUGCAUCAUCGGAUGCCUGUCCAAUUCGCGUCAGCACCCGAGUAUGCAGUUCUUUGCGUUUCCCCGGCCGGAGAACCCCUUCCACAAGCUGUGGAAGGAGGCGUGCCA ..((((((((((((((......((((....)))).......).))))))).(((.((((((....)))))))))....))))))...((((((((((.....)))))))).))....... ( -49.52, z-score = -2.45, R) >droWil1.scaffold_180698 8879812 120 - 11422946 CAUUCGGCGAUGCUGCAUUAUCGGCUGCCUGUCCAAUUCGCGACAGCAUCCGAGCAUGCAGUUCUUUGCGUUUCCCAGGCCAGAGAAUCCCUACCACAAGCUGUGGAAGGAGGCGUGCCA .....(((.(((((((((..((((.(((.(((((.....).))))))).))))..)))).((((((((.(((.....))))))))))).(((.((((.....)))).))).)))))))). ( -47.20, z-score = -2.47, R) >droVir3.scaffold_13049 16290418 120 + 25233164 UAUCCGGCGAUGCUGCAUCAUCGGAUGCUUGUCGAAUUCGCGGCAGCAUCCGAGCAUGCAGUUCUUCGCCUUUCCCAGGCCGGACAAUCCCUUUCACAAGCUCUGGAAAGAGGCGUGCCA .....(((((.(((((((..(((((((((.((((......)))))))))))))..)))))))...))))).....((.(((((.....))(((((.((.....))))))).))).))... ( -53.80, z-score = -4.15, R) >droMoj3.scaffold_6680 6403752 120 - 24764193 UAUACGGCGUUGCUGCAUCAUUGGAUGCUUGUCGAAUUCGAGGCAGCACCCAAACAUGCAGUUCUUUGCCUUUCCACGGCCUGACAAUCCCUUUCACAAGCUAUGGAAGGAGGCGUGCCA .....((((..(((((((..((((.(((((.(((....))).).)))).))))..))))))).....(((((((((..((.((.............)).))..))))..))))).)))). ( -42.92, z-score = -2.32, R) >droGri2.scaffold_15110 19859120 120 + 24565398 UAUACGGCGCUGUUGCAUCAUCGGAUGCCUCUCGAAUUCGCGGCAACAUCCGAGCAUGCAGUUCUUCGCUUUCCCCCGACCCGACUCCCCGUUCCACAAGCUGUGGAAGGAGGCGUGCCA .....(((((..((((((..(((((((((...((....)).))))...)))))..))))))......((((((........((......))((((((.....))))))))))))))))). ( -42.20, z-score = -1.66, R) >consensus CAUCCGGCGGUGCUGCAUCAUCGGAUGCCUGUCCAAUUCGCGGCAGCACCCGAGCAUGCAGUUCUUUGCGUUUCCCCGGCCGGAGAACCCCUUCCACAAGCUGUGGAAGGAGGCGUGCCA .....((((..(((((((..((((.(((.((.(........).))))).))))..))))))).........................(((.(((((.......))))).).))..)))). (-27.28 = -26.95 + -0.33)
| Location | 694,781 – 694,896 |
|---|---|
| Length | 115 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 76.67 |
| Shannon entropy | 0.48212 |
| G+C content | 0.57238 |
| Mean single sequence MFE | -39.79 |
| Consensus MFE | -19.82 |
| Energy contribution | -19.94 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.860915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 694781 115 + 24543557 GCCGGGGAAACGCAAAGAACUGCAUACUCGGGUGCUGACGCGAAUUGGACAAGCAUCCGAUGAUGCAGCACCGCCGGAUGGUCGGCAUCUCGACC-UGCGGAUGAGGAUGAAAUUG .(((.(....).)......((((((..(((((((((....(......)...)))))))))..))))))((((((.....((((((....))))))-.)))).)).))......... ( -41.90, z-score = -1.93, R) >droPer1.super_29 316510 101 - 1099123 GCCGUGGAAAGGCGAAGAACUGCAUGCUCGGAUGCUGCCGGGAAUUGGACAUGCAGCCGAUGAUGCAGCAACGCCGAAUGGUCGGCAUUCCGGAAACGUUG--------------- (((.......)))......((((((..((((.((((((((.....))).)).))).))))..))))))(((((((((....))))).....(....)))))--------------- ( -38.40, z-score = -1.60, R) >dp4.chrXR_group8 4666451 101 - 9212921 GCCGUGGAAAGGCGAAGAACUGCAUGCUCGGAUGCUGCCGGGAAUUGGACAUGCAGCCGAUGAUGCAGCAACGCCGAAUGGUCGGCAUUCCGGAAACGUUG--------------- (((.......)))......((((((..((((.((((((((.....))).)).))).))))..))))))(((((((((....))))).....(....)))))--------------- ( -38.40, z-score = -1.60, R) >droAna3.scaffold_13337 11638963 109 + 23293914 GACGAGGAAAGGCAAAGAACUGCAUGCUAGGAUGCUGGCGUGAGUUGGACAAACAGCCGAUGAUGCAGCAACGCCGAAUGGUCGGCAUCUCGGGC-UGCGGAAUUGCGUG------ .(((......(((....((((.((((((((....))))))))))))(......).)))(((..((((((...(((((....))))).......))-))))..))).))).------ ( -37.80, z-score = -1.27, R) >droEre2.scaffold_4784 704332 115 + 25762168 GGCGGGGGAACGCAAAGAACUGCAUACUAGGGUGCUGGCGCGAAUUGGACAGACAUCCGAUGAUGCAGCACCGCCGGAUGGUCGGCAUCUCGACC-UGCGGAUGCGGAUGGAAUUG .(((......)))......((((((.....(((((((.((...((((((......))))))..))))))))).(((.(.((((((....))))))-).)))))))))......... ( -44.40, z-score = -2.14, R) >droYak2.chr3L 684747 115 + 24197627 GGCGGGGAAACGCAAAGAACUGCAUGCUCGGGUGCUGGCGCGAAUUGGACAGACAUCCGAUGAUGCAGCACCGCCGGAUGGUCGGCAUCUCGACC-UGCGGAUGCGGAUGGAAUUG ((((((....)........((((((..((((((((((............))).)))))))..))))))..)))))...(.(((.(((((.((...-..))))))).))).)..... ( -47.50, z-score = -2.26, R) >droSec1.super_2 714779 97 + 7591821 GCCGGGGAAACGCAAAGAACUGCAUACUCGGGUGCUGACGCGAAUUGGACAGGCAUCCGAUGAUGCAGCACCGCCGGAUGGUCGGCAUCUCGAAU-UG------------------ ..(((((............((((((..(((((((((....(......)...)))))))))..))))))....(((((....))))).)))))...-..------------------ ( -35.10, z-score = -1.40, R) >droSim1.chr3L 344193 115 + 22553184 GCCGGGGAAACGCAAAGAACUGCAUACUCGGGUGCUGACGCGAAUUGGACAGGCAUCCGAUGAUGCAGCACCGCCGGAUGGUCGGCAUCUCGACC-UGCGGAUGCGGAUGGAAUUG .(((.(....).)......((((((.....(((((((.((...((((((......))))))..))))))))).(((.(.((((((....))))))-).)))))))))..))..... ( -45.10, z-score = -1.90, R) >droWil1.scaffold_180698 8879852 101 + 11422946 GCCUGGGAAACGCAAAGAACUGCAUGCUCGGAUGCUGUCGCGAAUUGGACAGGCAGCCGAUAAUGCAGCAUCGCCGAAUGGUCGGCAUUUCGGCAAAGUGG--------------- ((((((....).))..(((((((((..((((.(((((((........)))).))).))))..))))))....(((((....)))))..)))))).......--------------- ( -38.90, z-score = -1.85, R) >droVir3.scaffold_13049 16290458 98 - 25233164 GCCUGGGAAAGGCGAAGAACUGCAUGCUCGGAUGCUGCCGCGAAUUCGACAAGCAUCCGAUGAUGCAGCAUCGCCGGAUAGUCGGCAU---AACG-GGUAUA-------------- (((((...........((.((((((..(((((((((....((....))...)))))))))..))))))..))(((((....)))))..---..))-)))...-------------- ( -40.70, z-score = -2.78, R) >droMoj3.scaffold_6680 6403792 101 + 24764193 GCCGUGGAAAGGCAAAGAACUGCAUGUUUGGGUGCUGCCUCGAAUUCGACAAGCAUCCAAUGAUGCAGCAACGCCGUAUAGUCGGCAUGGUAACU-GGUAUA-------------- (((.......)))......((((((..(((((((((...(((....)))..)))))))))..))))))....((((......)))).........-......-------------- ( -37.10, z-score = -2.33, R) >droGri2.scaffold_15110 19859160 98 - 24565398 GUCGGGGGAAAGCGAAGAACUGCAUGCUCGGAUGUUGCCGCGAAUUCGAGAGGCAUCCGAUGAUGCAACAGCGCCGUAUAGUCGGCAU---AACG-UUUAUA-------------- .........(((((......(((((..(((((((((....((....))...)))))))))..))))).....((((......))))..---..))-)))...-------------- ( -32.20, z-score = -0.79, R) >consensus GCCGGGGAAACGCAAAGAACUGCAUGCUCGGAUGCUGCCGCGAAUUGGACAGGCAUCCGAUGAUGCAGCACCGCCGGAUGGUCGGCAUCUCGACC_UGUGGA______________ ...........(((.....((((((..((((.((...................)).))))..))))))....(((((....)))))..........)))................. (-19.82 = -19.94 + 0.11)
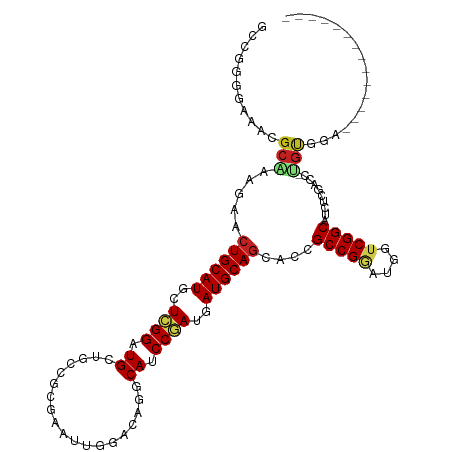
| Location | 694,781 – 694,896 |
|---|---|
| Length | 115 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 76.67 |
| Shannon entropy | 0.48212 |
| G+C content | 0.57238 |
| Mean single sequence MFE | -34.87 |
| Consensus MFE | -23.58 |
| Energy contribution | -23.55 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.980887 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
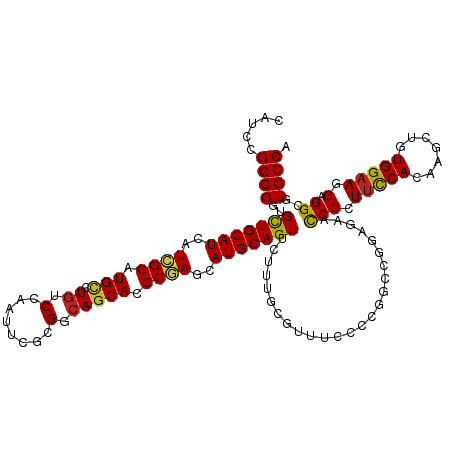
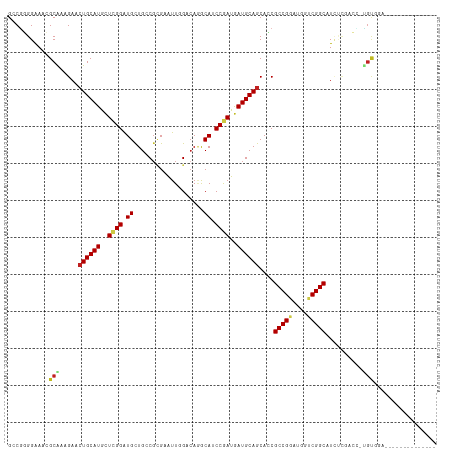
>dm3.chr3L 694781 115 - 24543557 CAAUUUCAUCCUCAUCCGCA-GGUCGAGAUGCCGACCAUCCGGCGGUGCUGCAUCAUCGGAUGCUUGUCCAAUUCGCGUCAGCACCCGAGUAUGCAGUUCUUUGCGUUUCCCCGGC ....................-(((((......)))))..((((.((.(((((((..((((.((((.(.((.....).).))))).))))..)))))))...........)))))). ( -35.70, z-score = -0.93, R) >droPer1.super_29 316510 101 + 1099123 ---------------CAACGUUUCCGGAAUGCCGACCAUUCGGCGUUGCUGCAUCAUCGGCUGCAUGUCCAAUUCCCGGCAGCAUCCGAGCAUGCAGUUCUUCGCCUUUCCACGGC ---------------........((((((((.....)))))((((..(((((((..((((.(((.((((........))))))).))))..)))))))....))))......))). ( -38.00, z-score = -2.84, R) >dp4.chrXR_group8 4666451 101 + 9212921 ---------------CAACGUUUCCGGAAUGCCGACCAUUCGGCGUUGCUGCAUCAUCGGCUGCAUGUCCAAUUCCCGGCAGCAUCCGAGCAUGCAGUUCUUCGCCUUUCCACGGC ---------------........((((((((.....)))))((((..(((((((..((((.(((.((((........))))))).))))..)))))))....))))......))). ( -38.00, z-score = -2.84, R) >droAna3.scaffold_13337 11638963 109 - 23293914 ------CACGCAAUUCCGCA-GCCCGAGAUGCCGACCAUUCGGCGUUGCUGCAUCAUCGGCUGUUUGUCCAACUCACGCCAGCAUCCUAGCAUGCAGUUCUUUGCCUUUCCUCGUC ------...((((....(((-((....((((((((....)))))))))))))......(((.((..(.....)..))))).((((......))))......))))........... ( -29.70, z-score = -1.13, R) >droEre2.scaffold_4784 704332 115 - 25762168 CAAUUCCAUCCGCAUCCGCA-GGUCGAGAUGCCGACCAUCCGGCGGUGCUGCAUCAUCGGAUGUCUGUCCAAUUCGCGCCAGCACCCUAGUAUGCAGUUCUUUGCGUUCCCCCGCC ...........((....)).-(((((......)))))....(((((((((((......((((....)))).......).)))))))...(.((((((....)))))).)....))) ( -33.12, z-score = -0.71, R) >droYak2.chr3L 684747 115 - 24197627 CAAUUCCAUCCGCAUCCGCA-GGUCGAGAUGCCGACCAUCCGGCGGUGCUGCAUCAUCGGAUGUCUGUCCAAUUCGCGCCAGCACCCGAGCAUGCAGUUCUUUGCGUUUCCCCGCC ..........((((.((((.-(((((......))))).....)))).(((((((..((((.((.(((.((.....).).))))).))))..)))))))....)))).......... ( -36.30, z-score = -0.81, R) >droSec1.super_2 714779 97 - 7591821 ------------------CA-AUUCGAGAUGCCGACCAUCCGGCGGUGCUGCAUCAUCGGAUGCCUGUCCAAUUCGCGUCAGCACCCGAGUAUGCAGUUCUUUGCGUUUCCCCGGC ------------------..-....(((((((((.((....))))(.(((((((..((((.(((.((.((.....).).))))).))))..))))))).)...)))))))...... ( -30.80, z-score = -0.72, R) >droSim1.chr3L 344193 115 - 22553184 CAAUUCCAUCCGCAUCCGCA-GGUCGAGAUGCCGACCAUCCGGCGGUGCUGCAUCAUCGGAUGCCUGUCCAAUUCGCGUCAGCACCCGAGUAUGCAGUUCUUUGCGUUUCCCCGGC ..........((((.((((.-(((((......))))).....)))).(((((((..((((.(((.((.((.....).).))))).))))..)))))))....)))).......... ( -37.70, z-score = -0.98, R) >droWil1.scaffold_180698 8879852 101 - 11422946 ---------------CCACUUUGCCGAAAUGCCGACCAUUCGGCGAUGCUGCAUUAUCGGCUGCCUGUCCAAUUCGCGACAGCAUCCGAGCAUGCAGUUCUUUGCGUUUCCCAGGC ---------------.......(((((((((((((....)))))((.(((((((..((((.(((.(((((.....).))))))).))))..))))))))).....)))))...))) ( -37.20, z-score = -2.79, R) >droVir3.scaffold_13049 16290458 98 + 25233164 --------------UAUACC-CGUU---AUGCCGACUAUCCGGCGAUGCUGCAUCAUCGGAUGCUUGUCGAAUUCGCGGCAGCAUCCGAGCAUGCAGUUCUUCGCCUUUCCCAGGC --------------......-....---..(((........(((((.(((((((..(((((((((.((((......)))))))))))))..)))))))...))))).......))) ( -43.76, z-score = -4.69, R) >droMoj3.scaffold_6680 6403792 101 - 24764193 --------------UAUACC-AGUUACCAUGCCGACUAUACGGCGUUGCUGCAUCAUUGGAUGCUUGUCGAAUUCGAGGCAGCACCCAAACAUGCAGUUCUUUGCCUUUCCACGGC --------------......-.........((((.......((((..(((((((..((((.(((((.(((....))).).)))).))))..)))))))....))))......)))) ( -32.82, z-score = -2.65, R) >droGri2.scaffold_15110 19859160 98 + 24565398 --------------UAUAAA-CGUU---AUGCCGACUAUACGGCGCUGUUGCAUCAUCGGAUGCCUCUCGAAUUCGCGGCAACAUCCGAGCAUGCAGUUCUUCGCUUUCCCCCGAC --------------......-((..---..((((......))))((((((((....(((((((((...((....)).))))...)))))))).)))))..............)).. ( -25.40, z-score = -0.55, R) >consensus ______________UCCACA_GGUCGAGAUGCCGACCAUCCGGCGGUGCUGCAUCAUCGGAUGCCUGUCCAAUUCGCGGCAGCACCCGAGCAUGCAGUUCUUUGCGUUUCCCCGGC .............................(((((......)))))..(((((((..((((.(((.((.(........).))))).))))..))))))).................. (-23.58 = -23.55 + -0.03)

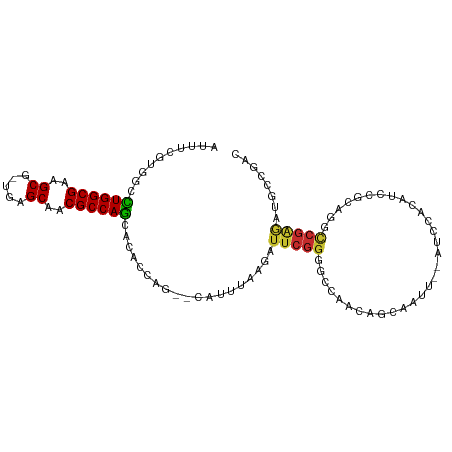
| Location | 694,861 – 694,961 |
|---|---|
| Length | 100 |
| Sequences | 9 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 68.60 |
| Shannon entropy | 0.62506 |
| G+C content | 0.53803 |
| Mean single sequence MFE | -30.86 |
| Consensus MFE | -13.86 |
| Energy contribution | -13.41 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.973122 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
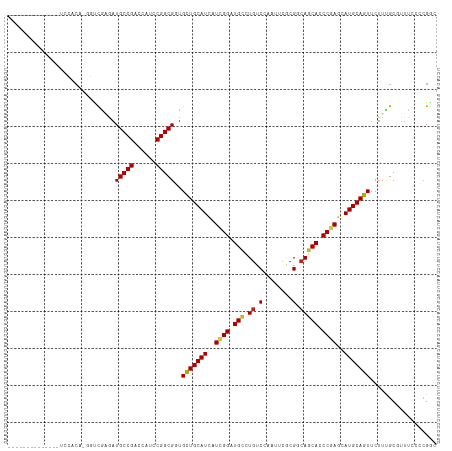
>dm3.chr3L 694861 100 - 24543557 AUUUCGUGACCUGGCGAAGCG-UGAGCAACGCCAGCACUCCAG--CAUUUAAGAUUCGGGGCCAACAGCAAUUUCAUCCUCAUCCGCAGGUCGAGAUGCCGAC .....(((..((((((..((.-...))..))))))))).....--..........((((.((.....)).(((((..((((....).)))..))))).)))). ( -30.30, z-score = -1.06, R) >droPer1.super_29 316590 90 + 1099123 AUUCGGUGGCUUGGCGGCGCAUUUAGCAACGCCAAGACAAA-AUUCAUUAAAAAUUCGGGGCAUACAG-------AACAACGUU-----UCCGGAAUGCCGAC ..(((((..(((((((..((.....))..))))))).....-...........((((.(((...((..-------......)).-----))).))))))))). ( -27.10, z-score = -2.33, R) >dp4.chrXR_group8 4666531 90 + 9212921 AUUCGGUGGCUUGGCGGCGCAUUUAGCAACGCCAAGACAAA-AUUCAUUAAAAAUUCGGGGCAUACAG-------AACAACGUU-----UCCGGAAUGCCGAC ..(((((..(((((((..((.....))..))))))).....-...........((((.(((...((..-------......)).-----))).))))))))). ( -27.10, z-score = -2.33, R) >droAna3.scaffold_13337 11639043 94 - 23293914 AUUCGUUGGCUUGGCGAAGCG-UCAGCAACGCCAGCUCCUAAA--GAUUUAAAAUUCGGGGCCAUCAGC------ACGCAAUUCCGCAGCCCGAGAUGCCGAC ....((((((((((((..((.-...))..))))))........--.........((((((((.....))------..((......))..))))))..)))))) ( -31.40, z-score = -1.42, R) >droEre2.scaffold_4784 704412 100 - 25762168 AUUUCGUGGCCUGGCGAAGCG-UGAGCAACGCCAGCACACCGG--CAUUUAAGAUUCGGGGCCAACAGCAAUUCCAUCCGCAUCCGCAGGUCGAGAUGCCGAC .....(((..((((((..((.-...))..))))))..)))(((--(((((..((((((((((.................)).))))..)))).)))))))).. ( -37.83, z-score = -2.14, R) >droYak2.chr3L 684827 100 - 24197627 AUUUCGUGGCCUGGCGAAGCG-UGAGCAACGCCAGCACACCAG--CAUUUAAGAUUCGGGGCCAACAGCAAUUCCAUCCGCAUCCGCAGGUCGAGAUGCCGAC ...(((((((((((((..((.-...))..)))).((......)--)............))))))...(((.(((.(((.((....)).))).))).)))))). ( -35.20, z-score = -1.92, R) >droSec1.super_2 714859 82 - 7591821 AUUUCGUGGCCUGGCGAAGCG-UGAGCAACGCCAGCACUCCAG--CAUUUAAGAUUCGGGGCCAACAGCAAUUCGAGAUGCCGAC------------------ ...(((((((((((((..((.-...))..)))).((......)--)............))))))...(((........)))))).------------------ ( -27.90, z-score = -1.54, R) >droSim1.chr3L 344273 100 - 22553184 AUUUCGUGGCCUGGCGAAGCG-UGAGCAACGCCAGCACUCCAG--CAUUUAAGAUUCGGGGCCAACAGCAAUUCCAUCCGCAUCCGCAGGUCGAGAUGCCGAC ...(((((((((((((..((.-...))..)))).((......)--)............))))))...(((.(((.(((.((....)).))).))).)))))). ( -35.20, z-score = -1.66, R) >droWil1.scaffold_180698 8879932 91 - 11422946 AAACGUCGUUUUGGCGGGGCAUUAAGCAACGCCAAAGCAAAGAUUCAUCCAGAAUUCGGAACAUAAAC-------AGCCACUUU-----GCCGAAAUGCCGAC ....((((.(((((((..((.....))..)))))))((((((((((.....))))..((.........-------..)).))))-----))........)))) ( -25.70, z-score = -2.18, R) >consensus AUUUCGUGGCCUGGCGAAGCG_UGAGCAACGCCAGCACACCAG__CAUUUAAGAUUCGGGGCCAACAGCAAUU__AUCCACAUCCGCAGGCCGAGAUGCCGAC ..........((((((..((.....))..))))))...................(((((...............................)))))........ (-13.86 = -13.41 + -0.44)

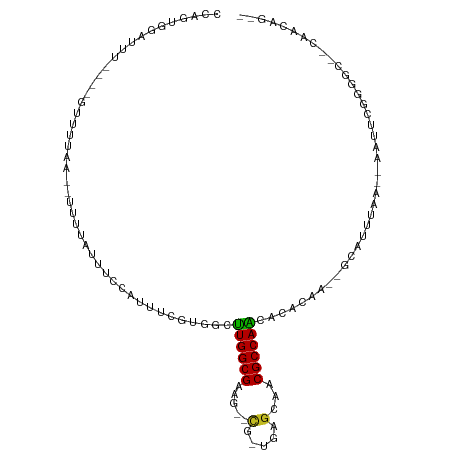
| Location | 694,896 – 694,989 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 64.12 |
| Shannon entropy | 0.72108 |
| G+C content | 0.43103 |
| Mean single sequence MFE | -25.38 |
| Consensus MFE | -8.34 |
| Energy contribution | -7.84 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.943674 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 694896 93 - 24543557 CCAGUGGAUUU----GUUUUAA--UUUCAUUUACAUUUCGUGACCUGGCGAAG--CG-UGAGCAACGCCAGCACUCCA--GCAUUUAA--GAUUCGGGGC--CAACAG-- ....(((....----(((((((--......((((.....)))).((((((..(--(.-...))..)))))).......--....))))--)))......)--))....-- ( -22.70, z-score = -0.84, R) >droPer1.super_29 316611 103 + 1099123 UUGGUGGACAUUGACGUUUAAUUCUUUUCUUUUCAUUCGGUGGCUUGGCGGCG--CAUUUAGCAACGCCAAGACAAAA-UUCAUUAAA--AAUUCGGGGC--AUACAGAA ((((((((((((((......................)))))).(((((((..(--(.....))..)))))))......-)))))))).--..........--........ ( -21.85, z-score = -0.44, R) >dp4.chrXR_group8 4666552 103 + 9212921 UUGGUGGACAUUGACGUUUAAUUCUUUUCUUUUCAUUCGGUGGCUUGGCGGCG--CAUUUAGCAACGCCAAGACAAAA-UUCAUUAAA--AAUUCGGGGC--AUACAGAA ((((((((((((((......................)))))).(((((((..(--(.....))..)))))))......-)))))))).--..........--........ ( -21.85, z-score = -0.44, R) >droAna3.scaffold_13337 11639072 93 - 23293914 CGGGUGGCCUU----UUUUGAU--UUUUUCUUUCAUUCGUUGGCUUGGCGAAG--CG-UCAGCAACGCCAGCUCCUAA--AGAUUUAA--AAUUCGGGGC--CAUCAG-- ..(((((((((----....(((--(((.(((((........((((.((((..(--(.-...))..))))))))...))--)))...))--)))).)))))--))))..-- ( -31.30, z-score = -2.65, R) >droEre2.scaffold_4784 704447 93 - 25762168 CCAGUGGAUUU----GUUUUAA--UUUUAUUUCCAUUUCGUGGCCUGGCGAAG--CG-UGAGCAACGCCAGCACACCG--GCAUUUAA--GAUUCGGGGC--CAACAG-- ..((((((..(----(......--...))..))))))...((((((((((..(--(.-...))..))))).....(((--(.......--...)))))))--))....-- ( -30.80, z-score = -2.55, R) >droYak2.chr3L 684862 93 - 24197627 CCAGUGGAUUU----GUUUUAA--UUUUAUUUCCAUUUCGUGGCCUGGCGAAG--CG-UGAGCAACGCCAGCACACCA--GCAUUUAA--GAUUCGGGGC--CAACAG-- ..((((((..(----(......--...))..))))))...((((((((((..(--(.-...))..)))).((......--))......--......))))--))....-- ( -30.20, z-score = -2.86, R) >droSec1.super_2 714876 93 - 7591821 CCAGUGGAUUU----GUUUUAA--UUUCAUUUACAUUUCGUGGCCUGGCGAAG--CG-UGAGCAACGCCAGCACUCCA--GCAUUUAA--GAUUCGGGGC--CAACAG-- ......((..(----((.....--........)))..)).((((((((((..(--(.-...))..)))).((......--))......--......))))--))....-- ( -27.22, z-score = -1.81, R) >droSim1.chr3L 344308 93 - 22553184 CCAGUGGAUUU----GUUUUAA--UUUCAUUUACAUUUCGUGGCCUGGCGAAG--CG-UGAGCAACGCCAGCACUCCA--GCAUUUAA--GAUUCGGGGC--CAACAG-- ......((..(----((.....--........)))..)).((((((((((..(--(.-...))..)))).((......--))......--......))))--))....-- ( -27.22, z-score = -1.81, R) >droWil1.scaffold_180698 8879953 90 - 11422946 --------UUU----UUUUUUGUUUUUUUUUAAAACGUCGUU--UUGGCGGGG--CAUUAAGCAACGCCAAAGCAAAGAUUCAUCCAG--AAUUCGGAACAUAAACAG-- --------...----.(((.((((((.............(((--((((((..(--(.....))..)))))))))...(((((.....)--)))).)))))).)))...-- ( -23.30, z-score = -3.52, R) >droVir3.scaffold_13049 16290556 86 + 25233164 --------UGC----AUUGAUAUUUUUAUGUUCUGAUUGGUA--UUGGCGGGC----AUUUGCAACGCCAAAAC-GA-CUUCAUUUAC--AAUUCGGGGCGUAAACAG-- --------...----.........(((((((((((((((((.--((((((.((----....))..)))))).))-(.-...).....)--)).))))))))))))...-- ( -24.70, z-score = -2.93, R) >droMoj3.scaffold_6680 6403893 89 - 24764193 --------AUU----GUUUAUAUAUUUAUGUUCUGAUUCGUA--UUGGCGGGCAUUUAUUUGCAACGCCAACAA-AA--UUCAUUAAC--AAUUCGGGGCGUAAACAG-- --------...----.........((((((((((((...((.--((((((.(((......)))..))))))...-..--.......))--...))))))))))))...-- ( -21.22, z-score = -2.65, R) >droGri2.scaffold_15110 19859258 99 + 24565398 --CCGAUUUUU----UUUUAUUUAUUUAUGUUCUAAUUGGUA-UUUGGCGGGCA-UUUUGUGCAACGCCAAAAACGA-CGAUAUUUGCAAAAUUCGUGGCGCCAAUAA-- --.........----....................((((((.-(((((((.(((-(...))))..)))))))..(.(-(((............)))).).))))))..-- ( -22.20, z-score = -0.66, R) >consensus CCAGUGGAUUU____GUUUUAA__UUUUAUUUCCAUUUCGUGGCUUGGCGAAG__CG_UGAGCAACGCCAACACACAA__GCAUUUAA__AAUUCGGGGC__CAACAG__ ............................................((((((...............))))))....................................... ( -8.34 = -7.84 + -0.49)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:53:56 2011