| Sequence ID | dm3.chr3L |
|---|---|
| Location | 691,349 – 691,473 |
| Length | 124 |
| Max. P | 0.999448 |

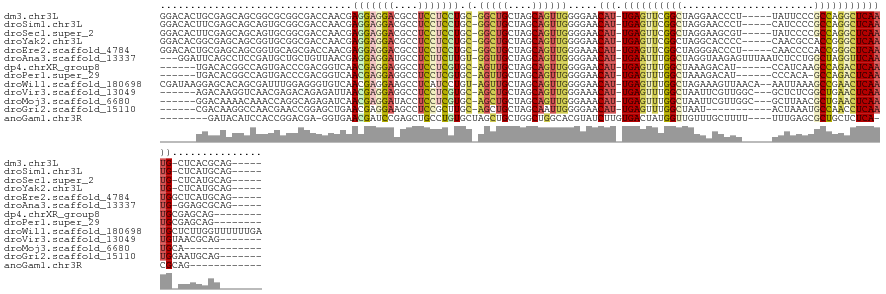
| Location | 691,349 – 691,473 |
|---|---|
| Length | 124 |
| Sequences | 13 |
| Columns | 137 |
| Reading direction | forward |
| Mean pairwise identity | 61.07 |
| Shannon entropy | 0.83214 |
| G+C content | 0.56061 |
| Mean single sequence MFE | -46.82 |
| Consensus MFE | -20.92 |
| Energy contribution | -20.55 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.56 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.90 |
| SVM RNA-class probability | 0.999448 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 691349 124 + 24543557 GGACACUGCGAGCAGCGGCGCGGCGACCAACGAGGAGGACGCCUCCUCCUGC-GGCUGCUAGCAGUUGGGGAACAU-UGAGUUCGGCUAGGAACCCU-----UAUUCCCGCCAGGCUCAAUG-CUCACGCAG----- ....(((((.((((((.((.((........))(((((((....)))))))))-.)))))).)))))((.(((.(((-(((((..(((..((((....-----..)))).)))..))))))))-.)).).)).----- ( -59.10, z-score = -3.31, R) >droSim1.chr3L 340774 124 + 22553184 GGACACUUCGAGCAGCAGUGCGGCGACCAACGAGGAGGACGCCUCCUCCUGC-GGCUGCUAGCAGUUGGGGAACAU-UGAGUUCGGCUAGGAACCCU-----CAUCCCCGCCAGGCUCAAUG-CUCAUGCAG----- ..........((((((...((((........(((((((...)))))))))))-.)))))).(((....(((..(((-(((((..(((..(((.....-----..)))..)))..))))))))-))).)))..----- ( -49.80, z-score = -1.20, R) >droSec1.super_2 711349 124 + 7591821 GGACACUUCGAGCAGCAGUGCGGCGACCAACGAGGAGGACGCCUCCUCCUGC-GGCUGCUAGCAGUUGGGGAACAU-UGAGUUCGGCUAGGAAGCGU-----UAUCCCCGCCAGGCUCAAUG-CUCAUGCAG----- ..........((((((...((((........(((((((...)))))))))))-.)))))).(((....(((..(((-(((((..(((..(((.....-----..)))..)))..))))))))-))).)))..----- ( -49.80, z-score = -1.06, R) >droYak2.chr3L 681030 124 + 24197627 GGACACGGCGAGCAGCGGUGCGGCGACCAACGAGGAGGACGCCUCCUCCUGC-GGCUGCUAGCAGUUGGGGAACAU-UGAGUUCGGCUAGGCACCCC-----CAACGCCACCGGGCUCAAUG-CUCAUGCAG----- .(((...((.(((((((((......)))..(((((((((....))))))).)-))))))).)).)))...((.(((-(((((((((...(((.....-----....))).))))))))))))-.))......----- ( -53.80, z-score = -1.18, R) >droEre2.scaffold_4784 700829 125 + 25762168 GGACACUGCGAGCAGCGGUGCAGCGACCAACGAGGAGGACGCCUCCUCCUGC-GGCUGCUAGCAGUUGGGAAACAU-UGAGUUCGGCUAGGGACCCU-----CAACCCCACCGGGCUCAAUGGCUCAUGCAG----- ....(((((.(((((((((......)))..(((((((((....))))))).)-))))))).)))))((((...(((-(((((((((...(((.....-----...)))..)))))))))))).)))).....----- ( -60.80, z-score = -3.63, R) >droAna3.scaffold_13337 11635504 126 + 23293914 ---GGAUUCAGCCUCCGAUGCUGCUGUUAACGAGGAGGAUGCCUCUUCUUGU-GGUUGCUAGCAGUUGGGGAACAU-UGAAUUUGGCUAGGUAAGAGUUUAAUCUCCUGGCUAGGUUCAAUG-GGAGCGCAG----- ---.......((((((...(((((((.(((((((((((...)))))))....-.)))).))))))).......(((-((((((((((((((..(((......))))))))))))))))))))-)))).))..----- ( -54.00, z-score = -4.16, R) >dp4.chrXR_group8 4662692 115 - 9212921 ------UGACACGGCCAGUGACCCGACGGUCAACGAGGAGGCCUCCUCGUGC-AGUUGCUAGCAGUUGGGGAACAU-UGAGUUUGGCUAAAGACAU------CCAUCAAGCCAGACUCAAUGCGAGCAG-------- ------.......(((.((..((((((.....(((((((....)))))))((-........)).))))))..))((-(((((((((((...((...------...)).)))))))))))))..).))..-------- ( -45.50, z-score = -3.06, R) >droPer1.super_29 312747 114 - 1099123 ------UGACACGGCCAGUGACCCGACGGUCAACGAGGAGGCCUCCUCGUGC-AGUUGCUAGCAGUUGGGGAACAU-UGAGUUUGGCUAAAGACAU------CCCACA-GCCAGACUCAAUGCGAGCAG-------- ------.......(((.((..((((((.....(((((((....)))))))((-........)).))))))..))((-(((((((((((........------.....)-))))))))))))..).))..-------- ( -45.32, z-score = -2.90, R) >droWil1.scaffold_180698 8875519 133 + 11422946 CGAUAAGGAGCACAGCGAUUUGGAGGGUGUCAACGAGGAAGCCUCAUCCUGU-AGUUGCUAGCAGUUGGGGAACAU-UGAGUUUGGCUAGAAAGUUAACA--AAUUAAAGCCGAACUCAAUGCUCUUGGUUUUUUGA ...((((((((..(((((((.(((((((.((......)).))))..)))...-)))))))........((((.(((-(((((((((((....((((....--))))..)))))))))))))).)))).)))))))). ( -43.50, z-score = -2.49, R) >droVir3.scaffold_13049 16287109 119 - 25233164 ------AGACAAGGUCAACGAGACAGAGAUUAACGAGGAGGCCUCCUCGUGC-AGCUGCUAGCAGUUGGGAAACAU-UGAGUUUGGCUAAUUCGUUGGC---GCUCUCGGCUGAACUCAAUGUAACGCAG------- ------.......(((.....)))........(((((((....))))))).(-(((((....))))))....((((-((((((..(((....((....)---).....)))..)))))))))).......------- ( -41.70, z-score = -1.81, R) >droMoj3.scaffold_6680 6400575 113 + 24764193 ------GGACAAAACAAACCAGGCAGAGAUCAACGAGGAUACCUCCUCGUGC-AGCUGCUAGCAGUUGGGAAACAU-UGAGUUUGGCUAAUUCGUUGGC---GCUUAACGCUGAACUCAAUGCA------------- ------............((............(((((((....))))))).(-(((((....))))))))...(((-((((((..((.....((....)---)......))..)))))))))..------------- ( -37.20, z-score = -2.28, R) >droGri2.scaffold_15110 19855025 111 - 24565398 ------CGACAAGGCCAACGAACCGGAGCUGAACGAGGAAGCCUCCGCUUGC-AGCUGCUAGCAAUUGGGGAACAU-UGAGUUUGGCUAAU-----------ACUAAAUGCCAACCUCAAUGGAAUGCAG------- ------........((((......((((((.........)).))))(((.((-....)).)))..))))....(((-((((.(((((....-----------.......))))).)))))))........------- ( -32.50, z-score = -1.16, R) >anoGam1.chr3R 31728265 111 + 53272125 --------GAUACAUCCACCGGACGA-GGUGAACGAUCCGAGCUGCCUGUGCUAGCUGCUGGCUGGCACGUAUCUUGUGACUAUGGUUGUUUGCUUUU----UUUGAGCGCUGCUCUCA-CGCAG------------ --------...........((((.((-((..(((((((((((.(((..((((((((.....))))))))))).)))).......)))))))..)))).----))))....((((.....-.))))------------ ( -35.61, z-score = -0.65, R) >consensus ______UGAGAACAGCAGUGCGGCGACGAUCAACGAGGACGCCUCCUCCUGC_AGCUGCUAGCAGUUGGGGAACAU_UGAGUUUGGCUAGGAACCUU_____CAUCCCCGCCAGGCUCAAUG_CUCACGC_______ ................................(((((((....)))))))...(((((....)))))......(((.((((((((((......................)))))))))))))............... (-20.92 = -20.55 + -0.37)
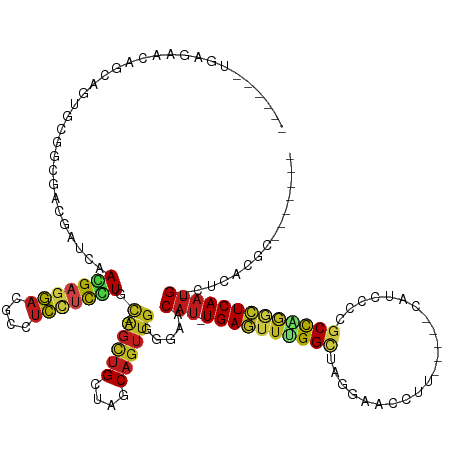
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:53:51 2011