| Sequence ID | dm3.chr3L |
|---|---|
| Location | 668,168 – 668,281 |
| Length | 113 |
| Max. P | 0.820831 |

| Location | 668,168 – 668,281 |
|---|---|
| Length | 113 |
| Sequences | 8 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 69.78 |
| Shannon entropy | 0.58267 |
| G+C content | 0.41980 |
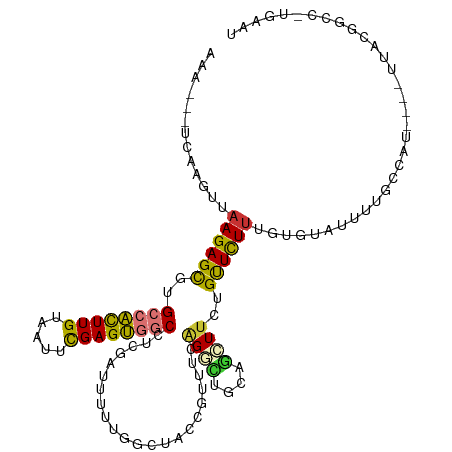
| Mean single sequence MFE | -28.89 |
| Consensus MFE | -13.39 |
| Energy contribution | -12.93 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.56 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.820831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
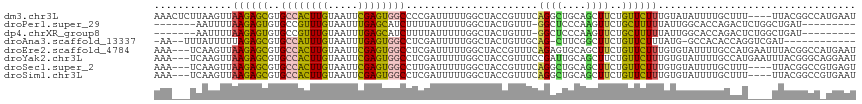
>dm3.chr3L 668168 113 + 24543557 AAACUCUUAAGUUAAGAGCGUGCCACUUGUAAUUCGAGUGGCCCCGAUUUUUGGCUACCGUUUCAGGCUGCAGCUUCUGUUCUUUGUAUAUUUUGCUUU----UUACGGCCAUGAAU .........(((((((((((.((((((((.....))))))))..)).))))))))).....(((((((((.(((....)))....(((.....)))...----...))))).)))). ( -30.70, z-score = -1.82, R) >droPer1.super_29 283983 100 - 1099123 -------AAUUUUAAGAGUGUGCCGUUUGUAAUUUGAGCAUCUUUUAUUUUUGGCUACUGUUU-GGCUCCCAAGUUCUGCUUUUUAUUGGCACCAGACUCUGGCUGAU--------- -------.....((((((.((((((.........)).))))))))))...(..((((..((((-((...((((.............))))..))))))..))))..).--------- ( -23.92, z-score = -1.08, R) >dp4.chrXR_group8 4637973 100 - 9212921 -------AAUUUUAAGAGUGUGCCGUUUGUAAUUUGAGCAUCUUUUAUUUUUGGCUACUGUUU-GGCUCCCAAGUUCUGCUUUUUAUUGGCACCAGACUCUGGCUGAU--------- -------.....((((((.((((((.........)).))))))))))...(..((((..((((-((...((((.............))))..))))))..))))..).--------- ( -23.92, z-score = -1.08, R) >droAna3.scaffold_13337 11611490 100 + 23293914 -AA--UUUAUUUUUAGAGCGUGCCAUUUGUAAUUUGAGUGGCCUCGAUUUUUGGCUACUGUUGCAG-CUUCGGCUUCUGUUCUUUAUG-GCCACACCAGGUCGAU------------ -..--.........((((((.(((..(((((((...(((((((.........))))))))))))))-....)))...))))))...((-(((......)))))..------------ ( -28.50, z-score = -1.86, R) >droEre2.scaffold_4784 676368 114 + 25762168 AAA---UCAAGUUAAGAGCGUGCCACUUGUAAUUCGAGUGGCCUCGAUUUUUGGCUACCGUUUCAGAGUGCAGCUUCUGUUCUUUGUGUAUUUUGCCAUGAAUUUACGGCCAUGAAU ...---(((.(((..(((...((((((((.....)))))))))))(((((.((((........(((((.((((...)))).)))))........)))).)))))...)))..))).. ( -31.99, z-score = -1.89, R) >droYak2.chr3L 656483 114 + 24197627 AAA---UCAAGUUAAGAGCGUGCCACUUGUAAUUCGAGUGGCCUCGAUUUUUGGCUACCGUUUCCGAUUGCAGCUUCUGUUCUUUGUGUAUUUUGCCAUGAAUUUACGGGCAGGAAU ...---...(((((((((((.((((((((.....))))))))..)).))))))))).....((((((..((((...))))))...........((((.((......)))))))))). ( -31.60, z-score = -1.35, R) >droSec1.super_2 688431 110 + 7591821 AAA---UCAAGUUAAGAGCGUGCCACUUGUAAUUCGAGUGGCCUUGAUUUUUGGCUACCGUUUCAGGCUGCAGCUUCUGUUCUUUGUGUAUUUUGCUUU----UUACGGCCGUGAGU ...---...(((((((((((.((((((((.....))))))))..)).))))))))).....(((((((((.(((....................)))..----...))))).)))). ( -29.15, z-score = -0.73, R) >droSim1.chr3L 317527 110 + 22553184 AAA---UCAAGUUAAGAGCGUGCCACUUGUAAUUCGAGUGGCCUCGAUUUUUGGCUACCGUUUCAGGCUGCAGCUUCUGUUCUUUGUGUAUUUUGCUUU----UUACGGCCGUGAAU ...---...(((((((((((.((((((((.....))))))))..)).))))))))).....(((((((((.(((....................)))..----...))))).)))). ( -31.35, z-score = -1.59, R) >consensus AAA___UCAAGUUAAGAGCGUGCCACUUGUAAUUCGAGUGGCCUCGAUUUUUGGCUACCGUUUCAGGCUGCAGCUUCUGUUCUUUGUGUAUUUUGCCAU____UUACGGCC_UGAAU .............((((((..((((((((.....))))))))......................((((....))))..))))))................................. (-13.39 = -12.93 + -0.47)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:53:47 2011