
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 663,920 – 663,995 |
| Length | 75 |
| Max. P | 0.995331 |

| Location | 663,920 – 663,995 |
|---|---|
| Length | 75 |
| Sequences | 10 |
| Columns | 79 |
| Reading direction | forward |
| Mean pairwise identity | 79.59 |
| Shannon entropy | 0.45363 |
| G+C content | 0.49143 |
| Mean single sequence MFE | -25.38 |
| Consensus MFE | -14.15 |
| Energy contribution | -14.40 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.53 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.79 |
| SVM RNA-class probability | 0.995331 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 663920 75 + 24543557 UUAACGCCCAGUCGGCGUUGCAAUUAUCCUUAUUAGUUGUGGCAUAAAACAUCGGUCC----CCUCGCCGAAGGGCGUA ...((((((..(((((((..((((((.......))))))..))..........((...----))..))))).)))))). ( -30.60, z-score = -4.33, R) >dp4.chrXR_group8 4633487 76 - 9212921 UUAACGCCCAGUCGGCGUUGCAAUUAUCCUUAUUAGUUGUGGCAUAAAGCCGAUGCCCGCGAUCUGGCCAAAGGGC--- .....((((.((((((((..((((((.......))))))..)).....))))))(((........)))....))))--- ( -27.60, z-score = -2.15, R) >droPer1.super_29 279366 79 - 1099123 UUAACGCCCAGUCGGCGUUGCAAUUAUCCUUAUUAGUUGUGGCAUAAAGCCGAUGCCCGCGAUCUGGCCAAAGGGCAUA .....((((.((((((((..((((((.......))))))..)).....))))))(((........)))....))))... ( -29.30, z-score = -2.48, R) >droSim1.chr3L 313336 75 + 22553184 UUAACGCCCAGUCGGCGUUGCAAUUAUCCUUAUUAGUUGUGGCAUAAAACAUCGGUCC----CCUCGCCAAAGGGCGUA ...((((((..(.(((((..((((((.......))))))..))..........((...----))..))))..)))))). ( -25.70, z-score = -3.15, R) >droSec1.super_2 684125 75 + 7591821 UUAACGCCCAGUCGGCGUUGCAAUUAUCCUUAUUAGUUGUGGCAUAAAACAUCGGUCC----CCUCGCCGAAGGGCGUA ...((((((..(((((((..((((((.......))))))..))..........((...----))..))))).)))))). ( -30.60, z-score = -4.33, R) >droYak2.chr3L 652142 75 + 24197627 UUAACGCCCAGUCGGCGUUGCAAUUAUCCUUAUUAGUUGUGGCAUAAAACAUCGGUCC----CCUUGCCGAAGGGCGUA ...((((((..(((((((..((((((.......))))))..))..........((...----))..))))).)))))). ( -30.30, z-score = -4.19, R) >droEre2.scaffold_4784 672063 75 + 25762168 UUAACGCCCAGUCGGCGCUGCAAUUAUCCUUAUUAGUUGUGGCAUAAAACAUCGGUCC----CCUUGCCGAAGGGCAUA .....((((..(((((((..((((((.......))))))..))..........((...----))..))))).))))... ( -28.60, z-score = -3.56, R) >droAna3.scaffold_13337 11607373 75 + 23293914 UUAACGCCCAGUCGGCGUUGCAAUUAUCCUUAUUAGUUGUGGCAUAAAACAUGGGUCC----CCUUACCAAAGGGUAUA .....(((((((....((..((((((.......))))))..)).....)).))))).(----((((....))))).... ( -21.30, z-score = -1.49, R) >droWil1.scaffold_180698 8841768 58 + 11422946 UUAACGCCCAGUCGGCGGCACAAUUAUCCUUAUUAGUUGUGGCAUAAAACAUCGCUUC--------------------- .....(((.....)))(.((((((((.......)))))))).)...............--------------------- ( -13.90, z-score = -1.95, R) >droMoj3.scaffold_6680 6371296 76 + 24764193 UUAACGCCCCGUCCGGCUAGCAAUUAUCCUUAUUAGUCCCGAC-CAUGGCAUAAAACACAUUUCUGGCCCAAAGGGC-- .....(((..(((.((((((.((......)).)))).)).)))-...)))................((((...))))-- ( -15.90, z-score = -0.08, R) >consensus UUAACGCCCAGUCGGCGUUGCAAUUAUCCUUAUUAGUUGUGGCAUAAAACAUCGGUCC____CCUCGCCAAAGGGCGUA .....(((.....)))((((((((((.......))))))))))......................((((....)))).. (-14.15 = -14.40 + 0.25)

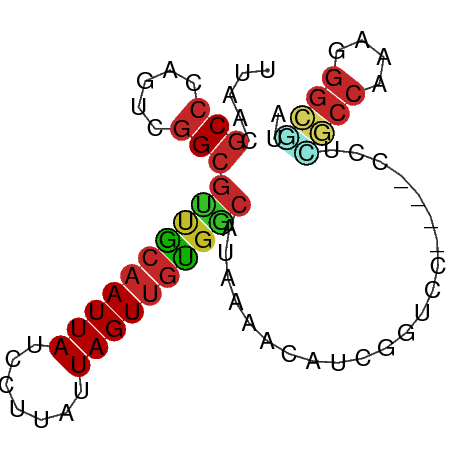
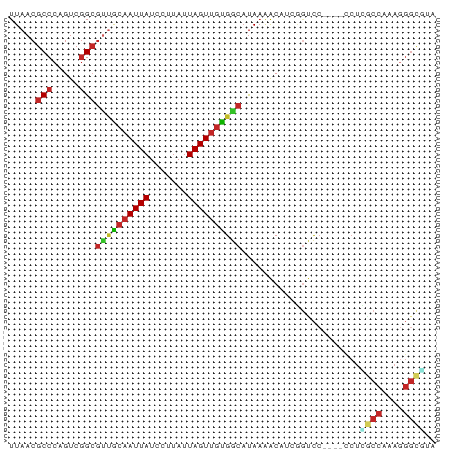
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:53:45 2011