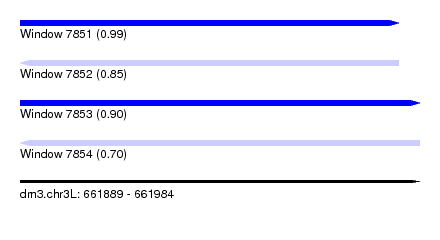
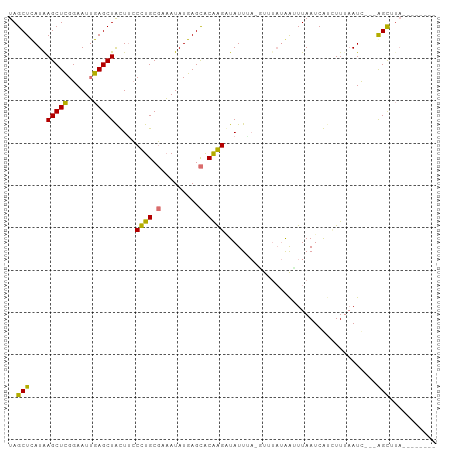
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 661,889 – 661,984 |
| Length | 95 |
| Max. P | 0.987595 |

| Location | 661,889 – 661,979 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 67.90 |
| Shannon entropy | 0.51764 |
| G+C content | 0.36720 |
| Mean single sequence MFE | -19.29 |
| Consensus MFE | -13.04 |
| Energy contribution | -14.22 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.28 |
| SVM RNA-class probability | 0.987595 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3L 661889 90 + 24543557 --------UAUGCU---GAUUAAAGACGAUUAAAUUAUACAUCUAAAUAACCCGUACUCAUAUUUAGCAGGUAAGUAGCUCAAUUCCGAGCUUAUAAGCUA --------..((((---((.((.(((((........................))).))..)).))))))(((....(((((......))))).....))). ( -13.96, z-score = -0.48, R) >droSim1.chr3L 311331 90 + 22553184 ACCCCCAAUAAGCU---GAUUAAAUACGAUUAAAUU--------AAAUACCCCGCGCUCAUAUUUCGCAGGGUAGCAGCUCAAUUCCGAGCUUAUAAGCUA ..........((((---...................--------...(((((.(((.........))).)))))..(((((......)))))....)))). ( -20.20, z-score = -2.33, R) >droYak2.chr3L 649996 88 + 24197627 -----------ACCCAUAAU--AAACUGAUUAAAUUAUAAACUUUAAUAUCUUGCGCUCAUAUUGCGCAAGGAAAUAGCUCAAUUCCGAGCUUAUGAGCUA -----------...(((((.--......((((((........)))))).((((((((.......)))))))).....((((......)))))))))..... ( -20.30, z-score = -3.01, R) >droEre2.scaffold_4784 670082 101 + 25762168 ACCCCGAAUAAGCUUGAGAUUAAAGAUGAUUAAGCCAUAAACGUCAAUAUCUUGUGCUCAUAUGGCGCAAGGAAAUAGCUCCAUUCCAAGCUCAUGAGCUA .....((((.((((..........((((.(((.....))).))))....(((((((((.....)))))))))....))))..))))..((((....)))). ( -22.70, z-score = -1.24, R) >consensus ________UAAGCU___GAUUAAAGACGAUUAAAUUAUAAAC_UAAAUAUCCCGCGCUCAUAUUGCGCAAGGAAAUAGCUCAAUUCCGAGCUUAUAAGCUA ...........((((..................................((((((((.......))))))))....(((((......)))))...)))).. (-13.04 = -14.22 + 1.19)

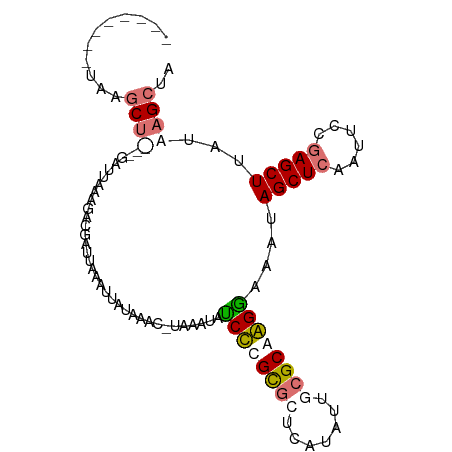
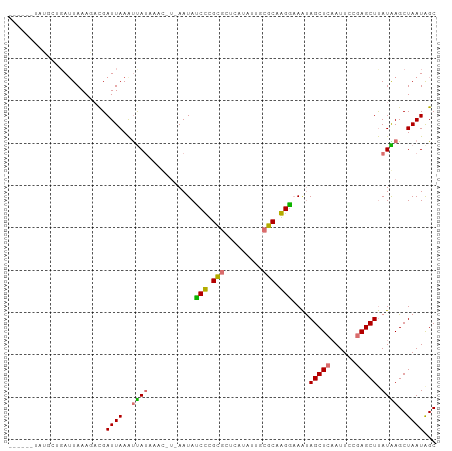
| Location | 661,889 – 661,979 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 67.90 |
| Shannon entropy | 0.51764 |
| G+C content | 0.36720 |
| Mean single sequence MFE | -22.85 |
| Consensus MFE | -14.04 |
| Energy contribution | -13.97 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.853955 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3L 661889 90 - 24543557 UAGCUUAUAAGCUCGGAAUUGAGCUACUUACCUGCUAAAUAUGAGUACGGGUUAUUUAGAUGUAUAAUUUAAUCGUCUUUAAUC---AGCAUA-------- ..(((.....(((((..(((.(((.........))).))).)))))...(((((...(((((...........))))).)))))---)))...-------- ( -16.80, z-score = -0.22, R) >droSim1.chr3L 311331 90 - 22553184 UAGCUUAUAAGCUCGGAAUUGAGCUGCUACCCUGCGAAAUAUGAGCGCGGGGUAUUU--------AAUUUAAUCGUAUUUAAUC---AGCUUAUUGGGGGU ...........(((.((..(((((((.(((((((((.........)))))))))...--------...((((......)))).)---)))))))).))).. ( -25.10, z-score = -1.18, R) >droYak2.chr3L 649996 88 - 24197627 UAGCUCAUAAGCUCGGAAUUGAGCUAUUUCCUUGCGCAAUAUGAGCGCAAGAUAUUAAAGUUUAUAAUUUAAUCAGUUU--AUUAUGGGU----------- ..((((((((.....(((((((........(((((((.......)))))))...(((((........))))))))))))--.))))))))----------- ( -24.00, z-score = -2.70, R) >droEre2.scaffold_4784 670082 101 - 25762168 UAGCUCAUGAGCUUGGAAUGGAGCUAUUUCCUUGCGCCAUAUGAGCACAAGAUAUUGACGUUUAUGGCUUAAUCAUCUUUAAUCUCAAGCUUAUUCGGGGU ..(((((((((((((((((((.((.........)).)))).(((((.((((((......)))).)))))))...........)).)))))))))...)))) ( -25.50, z-score = -0.72, R) >consensus UAGCUCAUAAGCUCGGAAUUGAGCUACUUCCCUGCGAAAUAUGAGCACAAGAUAUUUA_GUUUAUAAUUUAAUCAUCUUUAAUC___AGCUUA________ ..(((....(((((......))))).....(((((((.......)))))))....................................)))........... (-14.04 = -13.97 + -0.06)

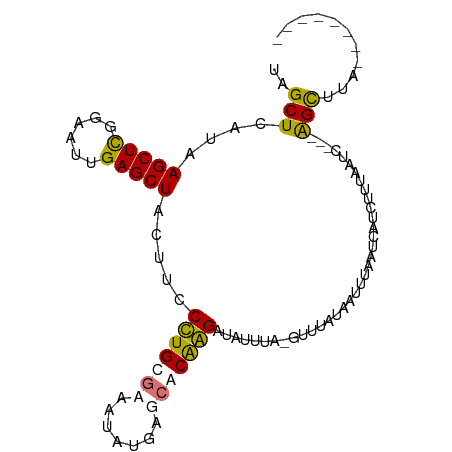
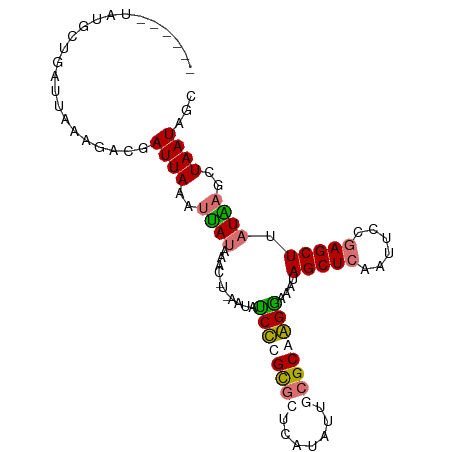
| Location | 661,889 – 661,984 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 70.37 |
| Shannon entropy | 0.48673 |
| G+C content | 0.35772 |
| Mean single sequence MFE | -19.59 |
| Consensus MFE | -13.54 |
| Energy contribution | -13.85 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.902879 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3L 661889 95 + 24543557 ------UAUGCUGAUUAAAGACGAUUAAAUUAUACAUCUAAAUAACCCGUACUCAUAUUUAGCAGGUAAGUAGCUCAAUUCCGAGCUUAUAAGCUAAUAGC ------..((((((.((.(((((........................))).))..)).))))))(((....(((((......))))).....)))...... ( -13.96, z-score = 0.05, R) >droSim1.chr3L 311336 90 + 22553184 ---CAAUAAGCUGAUUAAAUACGAUUAAAUUA--------AAUACCCCGCGCUCAUAUUUCGCAGGGUAGCAGCUCAAUUCCGAGCUUAUAAGCUAAUAGC ---.....((((....................--------..(((((.(((.........))).)))))..(((((......)))))....))))...... ( -20.20, z-score = -1.71, R) >droYak2.chr3L 650001 88 + 24197627 ------------UAAUAAAC-UGAUUAAAUUAUAAACUUUAAUAUCUUGCGCUCAUAUUGCGCAAGGAAAUAGCUCAAUUCCGAGCUUAUGAGCUAAUGGC ------------........-..((((..((((((.........((((((((.......)))))))).....((((......))))))))))..))))... ( -21.80, z-score = -2.57, R) >droEre2.scaffold_4784 670087 101 + 25762168 GAAUAAGCUUGAGAUUAAAGAUGAUUAAGCCAUAAACGUCAAUAUCUUGUGCUCAUAUGGCGCAAGGAAAUAGCUCCAUUCCAAGCUCAUGAGCUAAUAGC ((((.((((..........((((.(((.....))).))))....(((((((((.....)))))))))....))))..))))..((((....))))...... ( -22.40, z-score = -0.73, R) >consensus ______UAUGCUGAUUAAAGACGAUUAAAUUAUAAAC_U_AAUAUCCCGCGCUCAUAUUGCGCAAGGAAAUAGCUCAAUUCCGAGCUUAUAAGCUAAUAGC .......................((((..((((...........((((((((.......))))))))....(((((......))))).))))..))))... (-13.54 = -13.85 + 0.31)

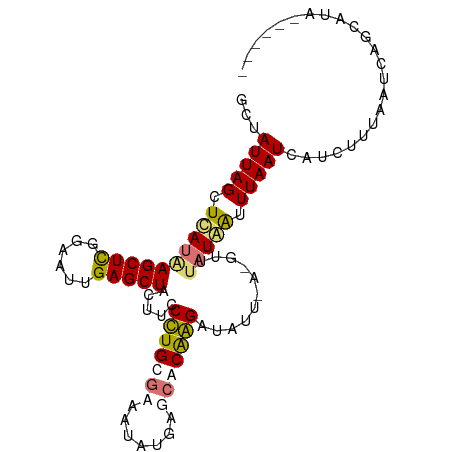
| Location | 661,889 – 661,984 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 70.37 |
| Shannon entropy | 0.48673 |
| G+C content | 0.35772 |
| Mean single sequence MFE | -21.91 |
| Consensus MFE | -13.30 |
| Energy contribution | -13.80 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.704448 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3L 661889 95 - 24543557 GCUAUUAGCUUAUAAGCUCGGAAUUGAGCUACUUACCUGCUAAAUAUGAGUACGGGUUAUUUAGAUGUAUAAUUUAAUCGUCUUUAAUCAGCAUA------ (((.(((((.....((((((....))))))........)))))...........(((((...(((((...........))))).))))))))...------ ( -19.22, z-score = -0.50, R) >droSim1.chr3L 311336 90 - 22553184 GCUAUUAGCUUAUAAGCUCGGAAUUGAGCUGCUACCCUGCGAAAUAUGAGCGCGGGGUAUU--------UAAUUUAAUCGUAUUUAAUCAGCUUAUUG--- ((((((((..(((.((((((....))))))..(((((((((.........)))))))))..--------..........))).))))).)))......--- ( -24.70, z-score = -1.67, R) >droYak2.chr3L 650001 88 - 24197627 GCCAUUAGCUCAUAAGCUCGGAAUUGAGCUAUUUCCUUGCGCAAUAUGAGCGCAAGAUAUUAAAGUUUAUAAUUUAAUCA-GUUUAUUA------------ .....(((((((............)))))))....(((((((.......)))))))..((((((........))))))..-........------------ ( -19.80, z-score = -1.45, R) >droEre2.scaffold_4784 670087 101 - 25762168 GCUAUUAGCUCAUGAGCUUGGAAUGGAGCUAUUUCCUUGCGCCAUAUGAGCACAAGAUAUUGACGUUUAUGGCUUAAUCAUCUUUAAUCUCAAGCUUAUUC ((.....))..(((((((((((((((.((.........)).)))).(((((.((((((......)))).)))))))...........)).))))))))).. ( -23.90, z-score = -0.86, R) >consensus GCUAUUAGCUCAUAAGCUCGGAAUUGAGCUACUUCCCUGCGAAAUAUGAGCACAAGAUAUU_A_GUUUAUAAUUUAAUCAUCUUUAAUCAGCAUA______ ...(((((.((((((((((......))))).....(((((((.......)))))))...........))))).)))))....................... (-13.30 = -13.80 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:53:45 2011