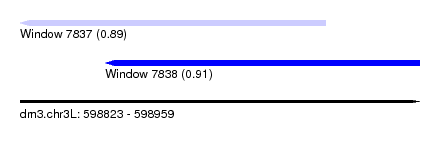
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 598,823 – 598,959 |
| Length | 136 |
| Max. P | 0.911608 |

| Location | 598,823 – 598,927 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 61.51 |
| Shannon entropy | 0.66500 |
| G+C content | 0.38120 |
| Mean single sequence MFE | -28.90 |
| Consensus MFE | -6.42 |
| Energy contribution | -5.65 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.57 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.22 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.885151 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 598823 104 - 24543557 ----UAUUUGGCAAUUCCAGUUACUUCUGGAUUUGCCGAAUACAAAUACA---AAUACAAA--CAUUAACUAAAUAGUGUAUCGUGGAGUGGAUCGAUUUCUUGGUAUUAGAA ----((((((((((.(((((......))))).))))))))))........---(((((..(--(((((......))))))..((.(((((......))))).))))))).... ( -28.40, z-score = -3.57, R) >droEre2.scaffold_4784 601050 103 - 25762168 ----UAUUUGACAAUUCCAGGCACUUGUGGAGUUGCCAAAUACAAAUUAA----AUACUAA--CAUUAACUAAAUAGUGUACCGUGGAGUGGAUCGAUUUCUUGGUAUGCAAA ----((((((.((((((((.(....).)))))))).))))))........----.......--.............((((((((.(((((......))))).))))))))... ( -26.40, z-score = -2.31, R) >droYak2.chr3L 581403 111 - 24197627 CACGUACUUGACAAUUCCAGUUACUUCUGGAGUUGCCAAGUACAAAUUAAUUAGAUACUAG--CAUUAACUAAAUAGUGUAUCGUGGAGUGGAUCGAUUUCUUGGUAUGUAAA ...(((((((.(((((((((......))))))))).))))))).......(((.(((((((--(((((......)))))).....(((((......))))).)))))).))). ( -34.80, z-score = -4.19, R) >droSim1.chr3L 248404 103 - 22553184 ----UAUUUGGCAACUCCAGUUGCUUUUGGAUUUGCCAAACACAAAU-UA---AAUGCUAA--CAUUAACUAAAUAGUGUAUCGUGGAGUGGAUCGAUUUCUUGGUAUUAAAA ----..((((((((.(((((......))))).)))))))).......-..---((((((((--(((((......)))))).....(((((......))))).))))))).... ( -25.60, z-score = -2.49, R) >dp4.chrXR_group3a 698996 99 + 1468910 ------UUCAAGGACACC-----CUUGUGCCUUGUGCGUACGCAGAUCUUCCUGGGAGCAGCCCGGGAAAUAAAUAUUAUUUUGCGAUGUGUAUAAACAGUUUCUCGUGC--- ------..(((((.(((.-----...))))))))...(((((.(((..((((((((.....))))))))..................(((......)))...))))))))--- ( -29.10, z-score = -1.68, R) >droPer1.super_23 860091 99 + 1662726 ------AUCAAGGACACC-----CUUGUGCCUUGUGCGUACGCAGAUCUUCCUGGGAGCAGCCCGGGAAAUAAAUAUUAUUUUGCGAUGUGUAUAAACAGUUUCUCGUGC--- ------..(((((.(((.-----...))))))))...(((((.(((..((((((((.....))))))))..................(((......)))...))))))))--- ( -29.10, z-score = -1.68, R) >consensus ____UAUUUGACAACUCCAGUUACUUGUGGAUUUGCCAAACACAAAUCUA___GAUACUAA__CAUUAACUAAAUAGUGUAUCGUGGAGUGGAUCGAUUUCUUGGUAUGCAAA ...........................(((.....)))........................................((((((.(((((......))))).))))))..... ( -6.42 = -5.65 + -0.77)
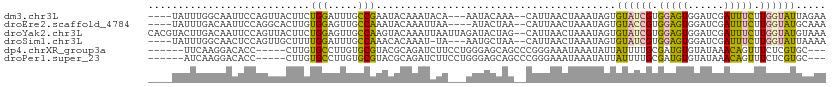
| Location | 598,852 – 598,959 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 53.95 |
| Shannon entropy | 0.80026 |
| G+C content | 0.37628 |
| Mean single sequence MFE | -28.24 |
| Consensus MFE | 0.00 |
| Energy contribution | 0.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 0.00 |
| Mean z-score | -3.78 |
| Structure conservation index | -0.00 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.911608 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 598852 107 - 24543557 ACUUUCCAUGGUACCCAGAAAGCUUAAAAUAUUAUUUGGCAAUUCCAGUUACUUCUG-GAUUUGC-CGAAUACAAAUACA---AAUACAAA--CAUUAACUAAAUAGUGUAUCG .(((((..(((...))))))))..........((((((((((.(((((......)))-)).))))-))))))........---.......(--(((((......)))))).... ( -27.30, z-score = -4.99, R) >droEre2.scaffold_4784 601079 90 - 25762168 ---------ACUUUCCAUUUAAUAU-------UAUUUGACAAUUCCAGGCACUUGUG-GAGUUGC-CAAAUACAAAUUAA----AUACUAA--CAUUAACUAAAUAGUGUACCG ---------.......(((((((..-------((((((.((((((((.(....).))-)))))).-))))))...)))))----))....(--(((((......)))))).... ( -19.70, z-score = -3.61, R) >droYak2.chr3L 581432 100 - 24197627 ---------ACUUUCAUUAUAAUAUCUCAC-GUACUUGACAAUUCCAGUUACUUCUG-GAGUUGC-CAAGUACAAAUUAAUUAGAUACUAG--CAUUAACUAAAUAGUGUAUCG ---------.....................-(((((((.(((((((((......)))-)))))).-)))))))..........(((((...--...............))))). ( -27.17, z-score = -5.28, R) >droSim1.chr3L 248433 106 - 22553184 ACUUUCCAUGGUGCCCAGAAAGCUUAAAAUAUUAUUUGGCAACUCCAGUUGCUUUUG-GAUUUGC-CAAACACAAAU-UA---AAUGCUAA--CAUUAACUAAAUAGUGUAUCG .(((((..(((...))))))))............((((((((.(((((......)))-)).))))-)))).......-..---.......(--(((((......)))))).... ( -22.90, z-score = -2.02, R) >dp4.chrXR_group3a 699022 103 + 1468910 -----------AUUUCCGUUCGCUCGCAGGAAUCUCCAGACUUCAAGGACACCCUUGUGCCUUGUGCGUACGCAGAUCUUCCUGGGAGCAGCCCGGGAAAUAAAUAUUAUUUUG -----------.((((((..(((((.((((((..((.......(((((.(((....))))))))(((....)))))..)))))).)))).)..))))))............... ( -36.20, z-score = -3.37, R) >droPer1.super_23 860117 103 + 1662726 -----------AUUUCCGUUCGCUCGCAGGAAUCUCCAGACAUCAAGGACACCCUUGUGCCUUGUGCGUACGCAGAUCUUCCUGGGAGCAGCCCGGGAAAUAAAUAUUAUUUUG -----------.((((((..(((((.((((((..((.......(((((.(((....))))))))(((....)))))..)))))).)))).)..))))))............... ( -36.20, z-score = -3.38, R) >consensus ___________UUUCCAGAUAGCUUAAAAGAAUAUUUGGCAAUUCCAGUCACUUUUG_GACUUGC_CAAACACAAAUCUA___GAUACUAA__CAUUAACUAAAUAGUGUAUCG .................................................................................................................. ( 0.00 = 0.00 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:53:31 2011