
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 583,289 – 583,367 |
| Length | 78 |
| Max. P | 0.990949 |

| Location | 583,289 – 583,367 |
|---|---|
| Length | 78 |
| Sequences | 12 |
| Columns | 87 |
| Reading direction | reverse |
| Mean pairwise identity | 78.28 |
| Shannon entropy | 0.44537 |
| G+C content | 0.43737 |
| Mean single sequence MFE | -22.37 |
| Consensus MFE | -15.60 |
| Energy contribution | -15.51 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.990949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 583289 78 - 24543557 -UUAAGGCAUUGAU----CGGCUUCCUGCUUCCGGCACGGAAGUCGUUCCAAA--ACGAGAGAAUUUCUAGUCUU-UAUAUUUAAA- -..(((((...((.----(((((((((((.....))).)))))))).))....--.(....)........)))))-..........- ( -21.60, z-score = -2.16, R) >droSim1.chr3L_random 27005 78 - 1049610 -UUAAGGCACCGAG----CGGCUUCCUGCUUCCGGCACGGAAGUCGUUCCAAA--ACGAGAGAAUUUCUAGUCUU-UACAUUUAAA- -..(((((...(((----(((((((((((.....))).)))))))))))....--.(....)........)))))-..........- ( -26.00, z-score = -3.12, R) >droSec1.super_2 603387 78 - 7591821 -UUAAGGCAUUGAG----CGGCUUCCUGCUUCCGGCACGGAAGUCGUUCCAAA--ACGAGAGAAUUUCUAGUCUU-UACAUUUAAA- -..(((((...(((----(((((((((((.....))).)))))))))))....--.(....)........)))))-..........- ( -25.70, z-score = -3.14, R) >droYak2.chr3L 565737 78 - 24197627 -UUAAGGCAUCGAU----CGACUUCCUGCUUCCGGCACGGAAGUCGCUCCCAA--ACGAGAGAAUUUCUAGUCUU-UACAUUUAAA- -..(((((...((.----(((((((((((.....))).)))))))).))....--.(....)........)))))-..........- ( -22.40, z-score = -2.59, R) >droEre2.scaffold_4784 585724 78 - 25762168 -UUAAGGCAUUGGU----CGGCUUCCUGCUUCCGGCACGGAAGUCGCUCCAAA--ACGAGAGAACUUCUAGUCUU-UACAUUUAAA- -..(((((.((((.----(((((((((((.....))).))))))))..)))).--.(....)........)))))-..........- ( -25.00, z-score = -2.75, R) >droAna3.scaffold_13337 21174657 82 + 23293914 -UUAAGGCAUUGGUG--GCGACUUCCUGCUUCCGGCACGGAAAUCGUUCCAAAACACGAGAGAUAUUCUAGUCUA-UAUCUUUAAA- -........((((.(--((((.(((((((.....))).)))).)))))))))......((((((((........)-)))))))...- ( -23.00, z-score = -2.36, R) >dp4.chrXR_group3a 682461 81 + 1468910 -UUAAGGCAUUGAGGAUGGGGCUUCCUGCUUCCCGCACGGAAGUCGGUCCAAA--ACGAGAGA-UAUCUAGUCUU-UAUCUUUAUA- -..(((((.....((((.((.((((((((.....))).))))))).))))...--...(((..-..))).)))))-..........- ( -23.20, z-score = -1.00, R) >droPer1.super_23 843446 81 + 1662726 -UUAAGGCAUUGAGGAUGGGGCUUCCUGCUUCCCGCACGGAAGUCGGUCCAAA--ACGAGAGA-UAUCUAGUCUU-UAUCUUUAAA- -((((((......((((.((.((((((((.....))).))))))).))))...--...(((((-(.....)))))-)..)))))).- ( -23.80, z-score = -1.19, R) >droWil1.scaffold_180698 1763206 67 + 11422946 UGUAAGGC----------CAAUUUGCGAUUUCCUGCACGGAAAUC----GA----ACGAGAGAAUCUCUAGUCUU-UAUCUUUAAA- ...(((((----------.......((((((((.....)))))))----).----..(((.....)))..)))))-..........- ( -16.20, z-score = -1.76, R) >droVir3.scaffold_13049 3427452 74 - 25233164 -UUAAGGCAUU------GCGAUUUCCUGCAU-CUGCACGGAAGUCCAA-GA----ACGAGCGAAUCUCUAGUCUUGUAUCUUUAAAA -.((((((.((------(.((((((((((..-..))).))))))))))-..----..(((.....)))..))))))........... ( -20.60, z-score = -2.32, R) >droMoj3.scaffold_6654 1978024 76 + 2564135 -GUAAGGCAUU------GCGAUUUCCUGCUUUCGGCACGGAAGUCCAAGAA----ACGAGCGAAUCUCUAGUCUUGUAUCUUUAGAG -.((((((.((------(.((((((((((.....))).))))))))))...----..(((.....)))..))))))........... ( -20.60, z-score = -1.11, R) >droGri2.scaffold_15110 7611094 71 - 24565398 -UUAAGGCAUU------GUGAUUUCCUGCAU-CUGCACGGAAGUC----CA----ACGAGCGAAUCUCUAGUCUUGUAUCUUUAACA -.((((((.((------(.((((((((((..-..))).)))))))----))----).(((.....)))..))))))........... ( -20.30, z-score = -2.70, R) >consensus _UUAAGGCAUUGA_____CGACUUCCUGCUUCCGGCACGGAAGUCGUUCCAAA__ACGAGAGAAUUUCUAGUCUU_UAUCUUUAAA_ ...(((((...........((((((((((.....))).)))))))............(((.....)))..)))))............ (-15.60 = -15.51 + -0.09)
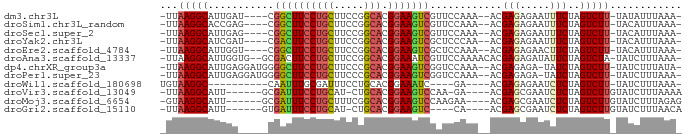
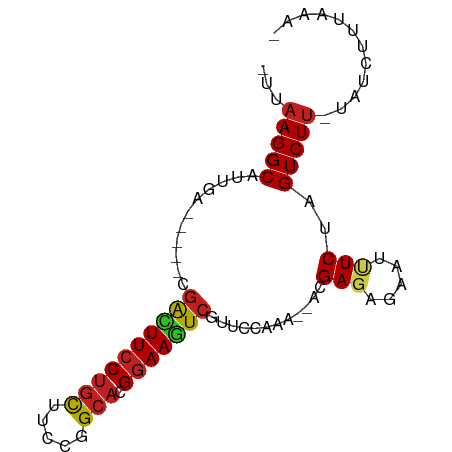
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:53:28 2011