| Sequence ID | dm3.chr3L |
|---|---|
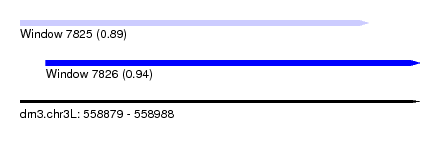
| Location | 558,879 – 558,988 |
| Length | 109 |
| Max. P | 0.940667 |

| Location | 558,879 – 558,974 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 83.35 |
| Shannon entropy | 0.30500 |
| G+C content | 0.42302 |
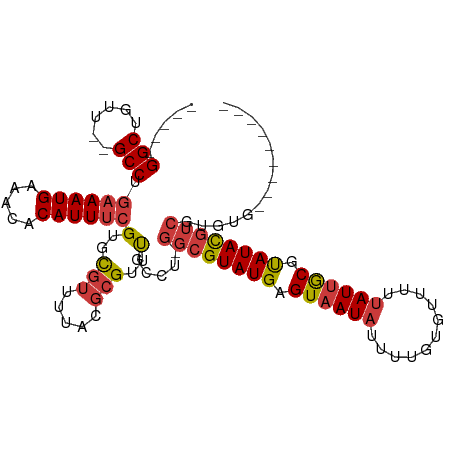
| Mean single sequence MFE | -30.11 |
| Consensus MFE | -20.77 |
| Energy contribution | -21.20 |
| Covariance contribution | 0.43 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.891022 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
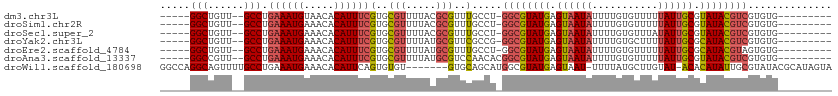
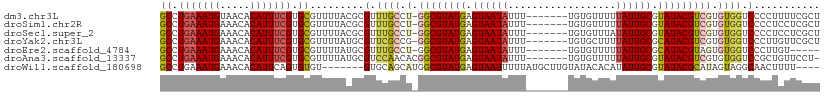
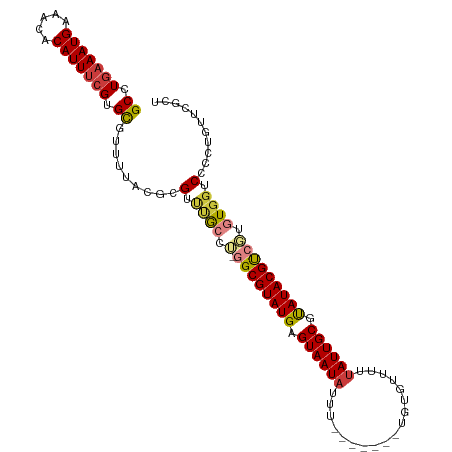
>dm3.chr3L 558879 95 + 24543557 -----GGCUGUU--GCCUGAAAUGUAACACAUUUCGUGCGUUUUACGCGUUUGCCU-GGCGUAUGAGUAAUAUUUUGUGUUUUUAUUGCGUAUACGUCGUGUG--------- -----(((....--))).((((((.....))))))..(((.....)))....((.(-((((((((.((((((...........)))))).))))))))).)).--------- ( -28.90, z-score = -2.21, R) >droSim1.chr2R 8780443 95 + 19596830 -----GGCUGUU--GCCUGAAAUGAAACACAUUUCGUGCGUUUUACGCGUUUGCCU-GGCGUAUGAGUAAUAUUUUGUGUUUUUAUUGCGUAUACGUCGUGUG--------- -----(((...(--((.(((((((...(((.....)))))))))).)))...))).-((((((((.((((((...........)))))).)))))))).....--------- ( -28.10, z-score = -1.94, R) >droSec1.super_2 579385 95 + 7591821 -----GGCUGUU--GCCUGAAAUGAAACACAUUUCGUGCGUUUUACGCGUUUGCCU-GGCGUAUGAGUAAUAUUUUGUGUUUAUAUUGCGUAUACGUCGUGUG--------- -----(((...(--((.(((((((...(((.....)))))))))).)))...))).-((((((((.(((((((.........))))))).)))))))).....--------- ( -29.10, z-score = -2.12, R) >droYak2.chr3L 540539 95 + 24197627 -----GGCUGUU--GCCUGAAAUGAAACACAUUUCGUGCGUUUUAUGCGUUCGCCG-GGCGUAUGAGUAAUAUUUUGUGCUUUUAUUGCGCAUACGUCGUGUG--------- -----(((...(--((.(((((((...(((.....)))))))))).)))...))).-((((((((.((((((...........)))))).)))))))).....--------- ( -30.20, z-score = -1.61, R) >droEre2.scaffold_4784 557689 95 + 25762168 -----GGCUGUU--GCCUGAAAUGAAACACAUUUCGUGCGUUUUAUGCGUUUGCCU-GGCGUAUGAGUAAUAUUUUGUGUUUUUAUUGCGCAUACGUAGUGUG--------- -----(((...(--((.(((((((...(((.....)))))))))).)))...))).-.(((((((.((((((...........)))))).)))))))......--------- ( -27.80, z-score = -1.78, R) >droAna3.scaffold_13337 21144936 96 - 23293914 -----GGCCGUU--GCCUGAAAUGAAACACAUUUCGUGCGUUUUAUGCGUCCAACACGGCGUAUGAGUAAUAUUUUGUGUUUUUAUUGCGUAUACGUCGUGUG--------- -----((.((((--((.(((((((.....))))))).)))......))).)).((((((((((((.((((((...........)))))).)))))))))))).--------- ( -33.70, z-score = -3.35, R) >droWil1.scaffold_180698 1729085 103 - 11422946 GGCCAGGCAGUUUUGCCUGAAAUGAAACACAUUCAGUGUGU-------GUGCAGCAUGGCGUAUGAGUAAU-UUUUAUGCUUGUAU-ACACAUAUUGCGUAUACGCAUAGUA ...(((((......)))))...((((.....))))((((((-------(((((....((((((.((....)-)..)))))))))))-))))))..((((....))))..... ( -33.00, z-score = -1.60, R) >consensus _____GGCUGUU__GCCUGAAAUGAAACACAUUUCGUGCGUUUUACGCGUUUGCCU_GGCGUAUGAGUAAUAUUUUGUGUUUUUAUUGCGUAUACGUCGUGUG_________ .....(((......))).((((((.....))))))(..(((.....)))..).....((((((((.((((((...........)))))).)))))))).............. (-20.77 = -21.20 + 0.43)
| Location | 558,886 – 558,988 |
|---|---|
| Length | 102 |
| Sequences | 7 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 81.96 |
| Shannon entropy | 0.34337 |
| G+C content | 0.43167 |
| Mean single sequence MFE | -29.46 |
| Consensus MFE | -19.82 |
| Energy contribution | -20.08 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.940667 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 558886 102 + 24543557 GCCUGAAAUGUAACACAUUUCGUGCGUUUUACGCGUUUGCCU-GGCGUAUGAGUAAUAUUU-------UGUGUUUUUAUUGCGUAUACGUCGUGUGGUCCCCUUUUCGCU ((.(((((((.....))))))).)).......(((.(..(.(-((((((((.((((((...-------........)))))).))))))))).)..).........))). ( -28.00, z-score = -2.19, R) >droSim1.chr2R 8780450 102 + 19596830 GCCUGAAAUGAAACACAUUUCGUGCGUUUUACGCGUUUGCCU-GGCGUAUGAGUAAUAUUU-------UGUGUUUUUAUUGCGUAUACGUCGUGUGGUCCCCUCCUCGCU ((..((((((.....))))))..(((.....)))....))..-((((((((.((((((...-------........)))))).))))))))(((.((......)).))). ( -27.80, z-score = -2.12, R) >droSec1.super_2 579392 102 + 7591821 GCCUGAAAUGAAACACAUUUCGUGCGUUUUACGCGUUUGCCU-GGCGUAUGAGUAAUAUUU-------UGUGUUUAUAUUGCGUAUACGUCGUGUGGUCCCCUCCUCGCU ((..((((((.....))))))..(((.....)))....))..-((((((((.(((((((..-------.......))))))).))))))))(((.((......)).))). ( -28.80, z-score = -2.31, R) >droYak2.chr3L 540546 102 + 24197627 GCCUGAAAUGAAACACAUUUCGUGCGUUUUAUGCGUUCGCCG-GGCGUAUGAGUAAUAUUU-------UGUGCUUUUAUUGCGCAUACGUCGUGUGGUCCCUUGUUCGCU ((.(((((((.....))))))).)).......(((..(((((-((((((((.((((((...-------........)))))).))))))))...)))).....)..))). ( -29.30, z-score = -1.10, R) >droEre2.scaffold_4784 557696 97 + 25762168 GCCUGAAAUGAAACACAUUUCGUGCGUUUUAUGCGUUUGCCU-GGCGUAUGAGUAAUAUUU-------UGUGUUUUUAUUGCGCAUACGUAGUGUGGUCCCUUGU----- ((.(((((((.....))))))).)).......(((...(((.-.(((((((.((((((...-------........)))))).))))))).....)))....)))----- ( -24.70, z-score = -1.13, R) >droAna3.scaffold_13337 21144943 102 - 23293914 GCCUGAAAUGAAACACAUUUCGUGCGUUUUAUGCGUCCAACACGGCGUAUGAGUAAUAUUU-------UGUGUUUUUAUUGCGUAUACGUCGUGUGGUCCGCUGUUCCU- ((.(((((((.....))))))).)).......(((.(((.(((((((((((.((((((...-------........)))))).))))))))))))))..))).......- ( -36.30, z-score = -4.26, R) >droWil1.scaffold_180698 1729099 99 - 11422946 GCCUGAAAUGAAACACAUUCAGUGUGU-------GUGCAGCAUGGCGUAUGAGUAAUUUUUAUGCUUGUAUACACAUAUUGCGUAUACGCAUAGUAGGCAACUUUU---- (((((...((((.....))))((((((-------(((((....((((((.((....))..)))))))))))))))))..((((....))))...))))).......---- ( -31.30, z-score = -2.06, R) >consensus GCCUGAAAUGAAACACAUUUCGUGCGUUUUACGCGUUUGCCU_GGCGUAUGAGUAAUAUUU_______UGUGUUUUUAUUGCGUAUACGUCGUGUGGUCCCCUGUUCGCU ((.(((((((.....))))))).))..................((((((((.((((((..................)))))).))))))))................... (-19.82 = -20.08 + 0.27)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:53:22 2011