
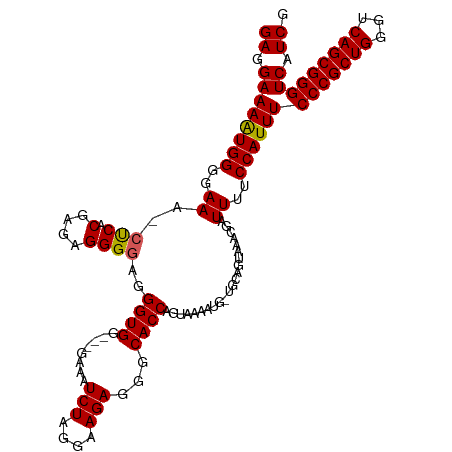
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 538,784 – 538,896 |
| Length | 112 |
| Max. P | 0.771696 |

| Location | 538,784 – 538,896 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 88.33 |
| Shannon entropy | 0.19191 |
| G+C content | 0.53405 |
| Mean single sequence MFE | -35.30 |
| Consensus MFE | -27.38 |
| Energy contribution | -27.26 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.575426 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 538784 112 + 24543557 GAGGAAAAUGGGGAAA-CCCACGAGAGGGGAGGGUGG---GAAAUCUAGGAAGAGGGCACCAGUAAAAUGUUGCAGUAAACGAUUUUCCAUUUCCCGCUGGGUCAGCGGGUCAUCG ........((((....-))))(((((.....((((((---((((((..........(((.((......)).))).(....)))))))))))))(((((((...))))))))).))) ( -37.10, z-score = -1.88, R) >droSim1.chr2R 8759622 112 + 19596830 GAGGAAAAUGGGGAAA-CUCACGAGAGGGGAGGGUGG---GAAAUCUAGGAAGAGGGCACCAGUAAAAUGCUGCAGUAAACGAUUUUCCAUUUCCCGCUGGGUCAGCGGGUCAUCC ..(((...((((....-)))).((.......((((((---((((((..........(((.((......)).))).(....)))))))))))))(((((((...))))))))).))) ( -34.40, z-score = -1.04, R) >droSec1.super_2 559494 112 + 7591821 GAGGAAAAUGGGGAAA-CUCACGAGAGGGGAGGGUGG---GAAAUCUAGGAAGAGGGCACCAGUAAAAUGUUGCAGUAAACGAUUUUCCAUUUCCCGCUGGGUCAGCGGGUCAUCG ........((((....-))))(((((.....((((((---((((((..........(((.((......)).))).(....)))))))))))))(((((((...))))))))).))) ( -34.10, z-score = -1.51, R) >droYak2.chr3L 519502 102 + 24197627 GAGGAAAGUGGGGAAA-CUCACGAGAGGGACGGGUGGUGGGAAAUCUAGGAAGAGGGCACC-------------AUUAAACGGUUUUCCAUUUCCCGCUGGGUCAGCGGGUCAUCG ((((((((((((....-)))))........((..(((((((...(((....)))...).))-------------))))..))..)))))....(((((((...)))))))...)). ( -38.60, z-score = -2.75, R) >droEre2.scaffold_4784 537371 100 + 25762168 GAGGAAAAUGGGGAAAACUCACGAGAGGGGUGGGUGG---GAAAUCUAGGAAGAGGGCACC-------------AUUAAACGGUUUUCCAUUUCCCGCUGGGUCAGCGGGUCAUCG (((((((((.(.....((((.(....))))).(((((---(...(((....)))...).))-------------)))...).)))))))....(((((((...)))))))...)). ( -32.30, z-score = -1.38, R) >consensus GAGGAAAAUGGGGAAA_CUCACGAGAGGGGAGGGUGG___GAAAUCUAGGAAGAGGGCACCAGUAAAAUG_UGCAGUAAACGAUUUUCCAUUUCCCGCUGGGUCAGCGGGUCAUCG ((.((((((((..((..(((.(....))))..((((........(((....)))...))))......................))..))))))(((((((...))))))))).)). (-27.38 = -27.26 + -0.12)


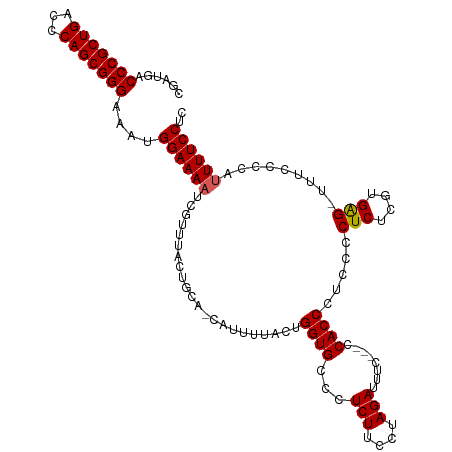
| Location | 538,784 – 538,896 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 88.33 |
| Shannon entropy | 0.19191 |
| G+C content | 0.53405 |
| Mean single sequence MFE | -26.58 |
| Consensus MFE | -20.92 |
| Energy contribution | -20.96 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.771696 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 538784 112 - 24543557 CGAUGACCCGCUGACCCAGCGGGAAAUGGAAAAUCGUUUACUGCAACAUUUUACUGGUGCCCUCUUCCUAGAUUUC---CCACCCUCCCCUCUCGUGGG-UUUCCCCAUUUUCCUC ......(((((((...)))))))....(((((((........(((.((......)).)))..........((...(---((((...........)))))-..))...))))))).. ( -27.70, z-score = -1.55, R) >droSim1.chr2R 8759622 112 - 19596830 GGAUGACCCGCUGACCCAGCGGGAAAUGGAAAAUCGUUUACUGCAGCAUUUUACUGGUGCCCUCUUCCUAGAUUUC---CCACCCUCCCCUCUCGUGAG-UUUCCCCAUUUUCCUC (((...(((((((...)))))))((((((.(((((((.....)).(((((.....)))))..........))))).---..........(((....)))-.....))))))))).. ( -27.80, z-score = -1.63, R) >droSec1.super_2 559494 112 - 7591821 CGAUGACCCGCUGACCCAGCGGGAAAUGGAAAAUCGUUUACUGCAACAUUUUACUGGUGCCCUCUUCCUAGAUUUC---CCACCCUCCCCUCUCGUGAG-UUUCCCCAUUUUCCUC ......(((((((...)))))))....(((((((.....................((((...(((....)))....---.)))).....(((....)))-.......))))))).. ( -24.40, z-score = -1.45, R) >droYak2.chr3L 519502 102 - 24197627 CGAUGACCCGCUGACCCAGCGGGAAAUGGAAAACCGUUUAAU-------------GGUGCCCUCUUCCUAGAUUUCCCACCACCCGUCCCUCUCGUGAG-UUUCCCCACUUUCCUC .((((.(((((((...)))))))((((((....))))))..(-------------((((...(((....))).....)))))..)))).(((....)))-................ ( -26.70, z-score = -2.22, R) >droEre2.scaffold_4784 537371 100 - 25762168 CGAUGACCCGCUGACCCAGCGGGAAAUGGAAAACCGUUUAAU-------------GGUGCCCUCUUCCUAGAUUUC---CCACCCACCCCUCUCGUGAGUUUUCCCCAUUUUCCUC .((((.(((((((...)))))))....(((((((........-------------((((...(((....)))....---.))))(((.......))).))))))).))))...... ( -26.30, z-score = -2.73, R) >consensus CGAUGACCCGCUGACCCAGCGGGAAAUGGAAAAUCGUUUACUGCA_CAUUUUACUGGUGCCCUCUUCCUAGAUUUC___CCACCCUCCCCUCUCGUGAG_UUUCCCCAUUUUCCUC ......(((((((...)))))))....((((((......................((((...(((....)))........)))).....(((....))).........)))))).. (-20.92 = -20.96 + 0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:53:18 2011