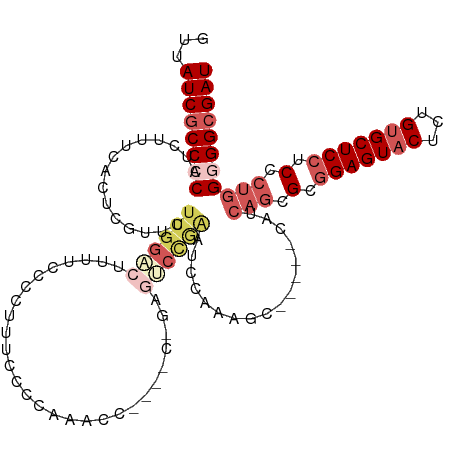
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 530,496 – 530,604 |
| Length | 108 |
| Max. P | 0.964530 |

| Location | 530,496 – 530,604 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.97 |
| Shannon entropy | 0.40080 |
| G+C content | 0.56724 |
| Mean single sequence MFE | -39.77 |
| Consensus MFE | -27.14 |
| Energy contribution | -26.68 |
| Covariance contribution | -0.46 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.896353 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 530496 108 + 24543557 AUCGCCCCCAGGGAGGAGCACAGAGUACUCCGCGCUGAUG-----GCU-UGGCUCUGGACUC------UGUUUGGGGAAAGAGGAAAAGUCCAAGAACGAGUGAAAGAUGGGGCGAUAAG (((((((((((((..((((.(((.((.....)).)))...-----)))-)..))))).((((------.((((..(((...........)))..)))))))).......))))))))... ( -41.30, z-score = -2.28, R) >droEre2.scaffold_4784 529091 120 + 25762168 AUCGCCUCCAGGGAGGAGCACAGAGUACUCUGCGUUGAUGGCUUGGCUUCGGCUUUGGCUUUGGAUUUGGUCUGGGGAAAGGGGAAAAGUCCAAGAACGAGUGAAAGAUGGGGCGAUAAC ((((((.(((.......((.((((....))))((((...((((.(((....)))..))))((((((((..(((........)))..)))))))).)))).))......)))))))))... ( -40.22, z-score = -1.28, R) >droYak2.chr3L 511030 110 + 24197627 AUCGCCUCCAGGGAGGAGCUCAGAGUACUCCGCGCUGAUG-----GCUUCGGAUUCAGGCUGGG-----GAAUGGGGAAUGGGGAAAAGCCCAAGAACGAGUGAAAGAUGGGGCGAUAAC ((((((.(((.......((((......((((((.((((..-----.(....)..)))))).)))-----).........((((......)))).....))))......)))))))))... ( -39.52, z-score = -1.67, R) >droSec1.super_2 551401 114 + 7591821 AUCGCCCCAGGGGAGGAGCACACAGUACUCCGCGCUGAUA-----GCUCUGGACUCGGACUCCGAAUCGGUUUGGGGAAAGAGGAAA-GUCCAAGAACGAGUGAGAGAUGGGGCGAUAAC ((((((((((.((((..((.....)).)))).).......-----.((((.(.((((..(((((((....))))))).....((...-..)).....))))).)))).)))))))))... ( -45.40, z-score = -2.82, R) >droSim1.chr2R 8750008 91 + 19596830 AUCCCCCCCAGGGAGGAGCACAGAGUACUCCGCGCUGAUG-----GCUCUGGACUCGGACUC------GGUUUGGGGAA------------------CGAGUGAAAGAUGGGGCGAUAAC (((((((((((((...(((.(.((....)).).)))....-----.))))))(((((..(((------......)))..------------------))))).......)))).)))... ( -32.40, z-score = -0.31, R) >consensus AUCGCCCCCAGGGAGGAGCACAGAGUACUCCGCGCUGAUG_____GCUCUGGAUUCGGACUC_G____GGUUUGGGGAAAGAGGAAAAGUCCAAGAACGAGUGAAAGAUGGGGCGAUAAC (((((((((((.(.((((.((...)).)))).).)))........((((....(((((((.........)))))))......((......))......)))).......))))))))... (-27.14 = -26.68 + -0.46)

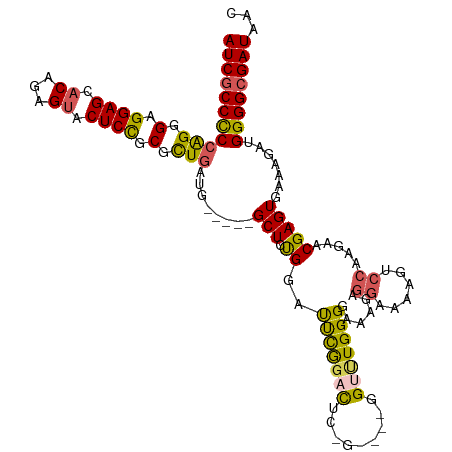
| Location | 530,496 – 530,604 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.97 |
| Shannon entropy | 0.40080 |
| G+C content | 0.56724 |
| Mean single sequence MFE | -33.74 |
| Consensus MFE | -20.12 |
| Energy contribution | -23.48 |
| Covariance contribution | 3.36 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.964530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 530496 108 - 24543557 CUUAUCGCCCCAUCUUUCACUCGUUCUUGGACUUUUCCUCUUUCCCCAAACA------GAGUCCAGAGCCA-AGC-----CAUCAGCGCGGAGUACUCUGUGCUCCUCCCUGGGGGCGAU ...((((((((...........(((((.(((((((................)------)))))))))))..-...-----...(((.(.(((((((...))))))).).))))))))))) ( -40.89, z-score = -4.46, R) >droEre2.scaffold_4784 529091 120 - 25762168 GUUAUCGCCCCAUCUUUCACUCGUUCUUGGACUUUUCCCCUUUCCCCAGACCAAAUCCAAAGCCAAAGCCGAAGCCAAGCCAUCAACGCAGAGUACUCUGUGCUCCUCCCUGGAGGCGAU ...(((((((((.............(((((.((((....((......))............((....)).))))))))).......((((((....))))))........))).)))))) ( -25.90, z-score = -1.25, R) >droYak2.chr3L 511030 110 - 24197627 GUUAUCGCCCCAUCUUUCACUCGUUCUUGGGCUUUUCCCCAUUCCCCAUUC-----CCCAGCCUGAAUCCGAAGC-----CAUCAGCGCGGAGUACUCUGAGCUCCUCCCUGGAGGCGAU ...(((((((((...............((((......))))..........-----....((((((.........-----..)))).))(((((.......)))))....))).)))))) ( -28.20, z-score = -0.73, R) >droSec1.super_2 551401 114 - 7591821 GUUAUCGCCCCAUCUCUCACUCGUUCUUGGAC-UUUCCUCUUUCCCCAAACCGAUUCGGAGUCCGAGUCCAGAGC-----UAUCAGCGCGGAGUACUGUGUGCUCCUCCCCUGGGGCGAU ...(((((((((..........(((((.((((-((..((((.((........))...))))...)))))))))))-----.........(((((((...))))))).....))))))))) ( -42.50, z-score = -3.64, R) >droSim1.chr2R 8750008 91 - 19596830 GUUAUCGCCCCAUCUUUCACUCG------------------UUCCCCAAACC------GAGUCCGAGUCCAGAGC-----CAUCAGCGCGGAGUACUCUGUGCUCCUCCCUGGGGGGGAU ...(((.((((.(((...(((((------------------..(.(......------).)..)))))..)))..-----...(((.(.(((((((...))))))).).))))))).))) ( -31.20, z-score = -1.25, R) >consensus GUUAUCGCCCCAUCUUUCACUCGUUCUUGGACUUUUCCCCUUUCCCCAAACC____C_GAGUCCGAAUCCAAAGC_____CAUCAGCGCGGAGUACUCUGUGCUCCUCCCUGGGGGCGAU ...((((((((...............((((((((........................)))))))).................(((.(.(((((((...))))))).).))))))))))) (-20.12 = -23.48 + 3.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:53:17 2011