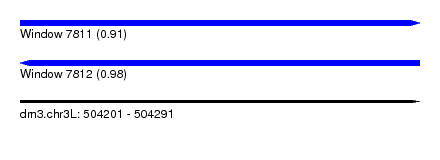
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 504,201 – 504,291 |
| Length | 90 |
| Max. P | 0.977579 |

| Location | 504,201 – 504,291 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.59 |
| Shannon entropy | 0.30431 |
| G+C content | 0.56696 |
| Mean single sequence MFE | -34.53 |
| Consensus MFE | -25.52 |
| Energy contribution | -25.90 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.912431 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
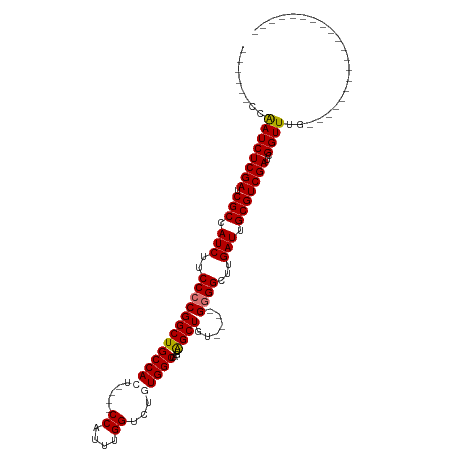
>dm3.chr3L 504201 90 + 24543557 ------CCAAUCUCGAUCGCCAUCUUCCCCGGCUGCCAAU----CCAUUUGGUCUGUGGCCAGUAGCUGU----GGGGCUUGAUUGCGUCGAUCGGUUUGUGUG---------------- ------.(((.(.((((((.(.....((((((((((....----((((.......))))...))))))).----)))((......))).))))))).)))....---------------- ( -30.80, z-score = -1.44, R) >droSim1.chr2R 8723128 86 + 19596830 ------CCAAUCUCGAUCGCCAUCUUCCCCGGCUGCCACU----CCAUUUGGUCUGUGGCCAGUAGCUGU----GGGGCUUGAUUGCGUCGAUCGGUUUG-------------------- ------..((((((((.(((.(((.((((((((((((((.----((....))...)))))....))))).----))))...))).)))))))..))))..-------------------- ( -33.20, z-score = -2.60, R) >droSec1.super_2 525709 86 + 7591821 ------CCAAUCUCGAUCGCCAUCUUCCCCGGCUGCCACU----CCAUUUGGUCUGUGGCCAGUAGCUUU----GGGGCUUGAUUGCGUCGAUCGGUUUG-------------------- ------..((((((((.(((.(((..(((((((((((((.----((....))...)))))....))))..----))))...))).)))))))..))))..-------------------- ( -31.90, z-score = -2.54, R) >droYak2.chr3L 482806 120 + 24197627 CCAAUCUCGAUCUCGAUCGCCAUCUUCCCCGGCUGCCAUUUGGACCACUUGGUCUUUGGCCAGUGGCUGUCUGUGCGGCUUGAUUGCGUCGAUCGGUUUGUGGAGUUGGAGUUGCAAUUG ((((.(((.((.((((((((((...((...((((((((..(((.(((((.(((.....)))))))))))..)).)))))).)).)).).)))))))...)).)))))))........... ( -42.20, z-score = -1.32, R) >consensus ______CCAAUCUCGAUCGCCAUCUUCCCCGGCUGCCACU____CCAUUUGGUCUGUGGCCAGUAGCUGU____GGGGCUUGAUUGCGUCGAUCGGUUUG____________________ ........((((((((.(((.(((..(((((((((((((.....((....))...)))))....))))......))))...))).)))))))..))))...................... (-25.52 = -25.90 + 0.38)

| Location | 504,201 – 504,291 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.59 |
| Shannon entropy | 0.30431 |
| G+C content | 0.56696 |
| Mean single sequence MFE | -31.40 |
| Consensus MFE | -24.61 |
| Energy contribution | -25.68 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.977579 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
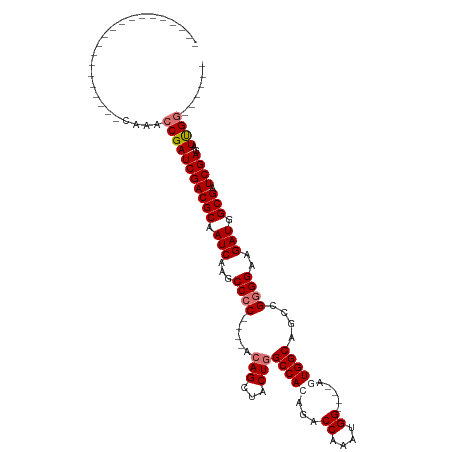
>dm3.chr3L 504201 90 - 24543557 ----------------CACACAAACCGAUCGACGCAAUCAAGCCCC----ACAGCUACUGGCCACAGACCAAAUGG----AUUGGCAGCCGGGGAAGAUGGCGAUCGAGAUUGG------ ----------------........(((((((((((.(((...((((----.(((...)))((((....((....))----..))))....))))..))).))).))))..))))------ ( -29.20, z-score = -2.52, R) >droSim1.chr2R 8723128 86 - 19596830 --------------------CAAACCGAUCGACGCAAUCAAGCCCC----ACAGCUACUGGCCACAGACCAAAUGG----AGUGGCAGCCGGGGAAGAUGGCGAUCGAGAUUGG------ --------------------....(((((((((((.(((...((((----.(((...)))(((((...((....))----.)))))....))))..))).))).))))..))))------ ( -32.40, z-score = -3.20, R) >droSec1.super_2 525709 86 - 7591821 --------------------CAAACCGAUCGACGCAAUCAAGCCCC----AAAGCUACUGGCCACAGACCAAAUGG----AGUGGCAGCCGGGGAAGAUGGCGAUCGAGAUUGG------ --------------------....(((((((((((.(((...((((----...(((....(((((...((....))----.)))))))).))))..))).))).))))..))))------ ( -31.70, z-score = -3.07, R) >droYak2.chr3L 482806 120 - 24197627 CAAUUGCAACUCCAACUCCACAAACCGAUCGACGCAAUCAAGCCGCACAGACAGCCACUGGCCAAAGACCAAGUGGUCCAAAUGGCAGCCGGGGAAGAUGGCGAUCGAGAUCGAGAUUGG ........................((((((..(((.(((...((.(.(((.......)))((((..((((....))))....))))....).))..))).))).(((....))))))))) ( -32.30, z-score = -0.45, R) >consensus ____________________CAAACCGAUCGACGCAAUCAAGCCCC____ACAGCUACUGGCCACAGACCAAAUGG____AGUGGCAGCCGGGGAAGAUGGCGAUCGAGAUUGG______ ........................(((((((((((.(((...((((.....(((...)))(((((...((....)).....)))))....))))..))).))).))))..))))...... (-24.61 = -25.68 + 1.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:53:10 2011