| Sequence ID | dm3.chr3L |
|---|---|
| Location | 501,801 – 501,899 |
| Length | 98 |
| Max. P | 0.737351 |

| Location | 501,801 – 501,899 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 57.61 |
| Shannon entropy | 0.78925 |
| G+C content | 0.48683 |
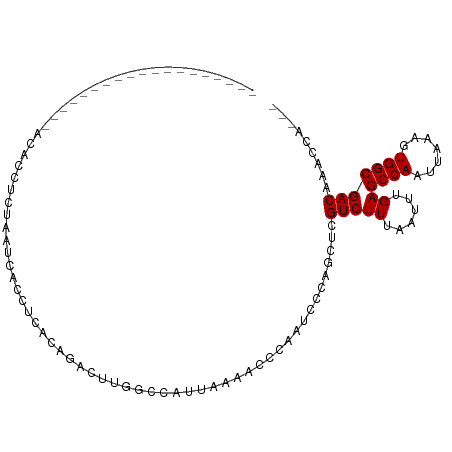
| Mean single sequence MFE | -17.10 |
| Consensus MFE | -5.54 |
| Energy contribution | -5.54 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.737351 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 501801 98 + 24543557 --------CUCCCCCCCUCCCACCACCCCUUACCCGACAGACGUUGCCAUUAAAACCCAAUCCCAGCUCGUCUUUAAUUUGAGCCAAUUAAAGUGGCGACAAAGCA--- --------..................................(((((((((..............(((((.........))))).......)))))))))......--- ( -14.70, z-score = -2.18, R) >droSim1.chr2R 8720772 89 + 19596830 -----------------GAACCCCCCUCACCACCUCACAGAGAUUGCCAUUAAAACCCAAUCCCAGCUCGUCUUUAAUUUGAGCCAAUUAAAGUGGCGACAAAGCA--- -----------------.........((.((((......(.(((((...........))))).).(((((.........)))))........)))).)).......--- ( -14.10, z-score = -1.95, R) >droSec1.super_2 523524 83 + 7591821 -------------------ACACUUCUAAACACCUCGCAGAGAUUGCCAUUAAAACCCAAUCCCAGCUCGUCUUUAAUUUGAGCCAAUUAAAGUGGCGACAA------- -------------------...............((((.(.(((((...........))))).).(((((.........)))))...........))))...------- ( -12.60, z-score = -1.03, R) >droEre2.scaffold_4784 499731 80 + 25762168 -------------------------UCCAGCUCCUCCCCGGCGUGUCCAU-AAAACCCAAUCCCAGCUCGUCUUUAAUUUGAGCCAAUUAAAGUGGCGACAAAGCA--- -------------------------....(((..((.((((.((......-...)))).......(((((.........)))))..........)).))...))).--- ( -13.00, z-score = -0.47, R) >droWil1.scaffold_180727 2425877 92 - 2741493 --------------AGAUGGAAACUCUAACUAACUAACCAACUCCUCGA--GAGAUUUAGAGUUGGAGCGUCUUUAAUUUGAGCCAAUUAAA-UGGCGACAAUCCAAUC --------------...((((................(((((((.((..--..))....))))))).((((((((((((......)))))))-.)))).)..))))... ( -22.90, z-score = -2.48, R) >droMoj3.scaffold_6654 1879715 100 - 2564135 CCCCCUCCUCCUUCUC----CUCCUCUACUCUGUCUCUACUCUUGGCAAUUGGCACUCAUCCGAGUCGCGUCUUGAAUUUGAGCCAAUUAAA-UGGCGACACUCG---- ................----...........((((.(((...(((((.....((((((....)))).)).((........))))))).....-))).))))....---- ( -17.50, z-score = -1.28, R) >droVir3.scaffold_13049 3300374 103 + 25233164 CCCCGCACCCCGCCGCGUAACUCCACAACUCCA-CGCUGGGCUAGGCAAUUGGCACUCAUCCGAGUCGCGUCUUGAAUUUGAGCCAAUUAAA-UGGCGACACUCG---- ..........((((((.((.(.((((.......-.).)))).)).))(((((((.(..((.((((......)))).))..).)))))))...-.)))).......---- ( -24.90, z-score = 0.19, R) >consensus ___________________ACACCUCUAAUCACCUCACAGACUUGGCCAUUAAAACCCAAUCCCAGCUCGUCUUUAAUUUGAGCCAAUUAAAGUGGCGACAAACCA___ .....................................................................(((((......))((((.......)))))))......... ( -5.54 = -5.54 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:53:09 2011