
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 496,969 – 497,130 |
| Length | 161 |
| Max. P | 0.885751 |

| Location | 496,969 – 497,063 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 81.84 |
| Shannon entropy | 0.32709 |
| G+C content | 0.49473 |
| Mean single sequence MFE | -30.12 |
| Consensus MFE | -18.50 |
| Energy contribution | -19.95 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.636250 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
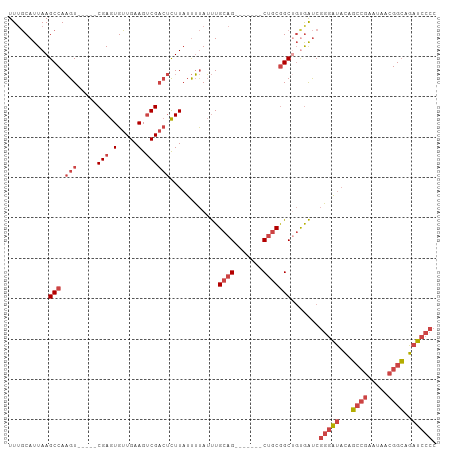
>dm3.chr3L 496969 94 + 24543557 UUUGCAUUAAGCCAAGU-----CGAGUGUUGAAGUCGACUCUUAUUUUAUUUGCAG-------CUGCGGCUGCGAUCGGGAUACAGCCGAAUAACGGCAGAUCCCC .......((((...(((-----(((.(.....).))))))))))......((((((-------(....)))))))..(((((...((((.....))))..))))). ( -32.70, z-score = -2.78, R) >droPer1.super_23 713965 105 - 1662726 UUUACAUUAAGCCACGUUGUGUCGAGUGUUGAAGCCGCAAUUUCUCGCAGCUGUAGGCCUCUGCCUCAGCCUCAGCCUCUGCCUCUCCACAUAACGGAGGAUCCC- ...........((.((((((((.(((.((.((.((.((........)).((((.((((....))))))))....)).)).))..))).))))))))..)).....- ( -33.20, z-score = -2.29, R) >droSim1.chr2R 8715679 94 + 19596830 UUUGCAUUAAGCCAAGU-----CGAGUGUUGAAGUCGACUCUUAUUUUAUUUGCAG-------CUGCGGCUGUGAUCGGGAUACAGCCGAAUAACGGCAGAUCCCC .......((((...(((-----(((.(.....).))))))))))......(..(((-------(....))))..)..(((((...((((.....))))..))))). ( -30.50, z-score = -2.31, R) >droSec1.super_2 518757 94 + 7591821 UUUGCAUUAAGCCAAGU-----CGAGUGUUGAAGUCGACUCUUAUUUUAUUUGCAG-------CUGCGGCUGUGAUCGGGAUACAGCCGAAUAACGGCAGAUCCCC .......((((...(((-----(((.(.....).))))))))))......(..(((-------(....))))..)..(((((...((((.....))))..))))). ( -30.50, z-score = -2.31, R) >droYak2.chr3L 475482 94 + 24197627 UUGGCAUUAAGCCAAGU-----CGAGUGUUGAAGUCGACUCUUAUUUUAUUUGCAG-------CUGCGGCGGUGAUCGGGAUACAGCGGAAUAACGGCAGAUCCCC (((((.....)))))((-----(((.(.....).)))))...........(..(.(-------(....)).)..)..(((((...((.(.....).))..))))). ( -25.90, z-score = -0.17, R) >droEre2.scaffold_4784 494780 94 + 25762168 UUUGCAUUAAGCCAAGU-----CGAGUGUUGAAGUCGACUCUUAUUUUAUUUGCAG-------CUGCGGCUGUGAUCGGGAUACAGGCGAAUAACGGCAGAUCCCC (((((.....(((.(((-----(((.(.....).))))))..((((((..(..(((-------(....))))..)..))))))..)))........)))))..... ( -27.92, z-score = -1.24, R) >consensus UUUGCAUUAAGCCAAGU_____CGAGUGUUGAAGUCGACUCUUAUUUUAUUUGCAG_______CUGCGGCUGUGAUCGGGAUACAGCCGAAUAACGGCAGAUCCCC ..........(((..........(((.((((....)))).))).........((((.......))))))).......(((((...((((.....))))..))))). (-18.50 = -19.95 + 1.45)


| Location | 496,969 – 497,063 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 81.84 |
| Shannon entropy | 0.32709 |
| G+C content | 0.49473 |
| Mean single sequence MFE | -29.35 |
| Consensus MFE | -17.06 |
| Energy contribution | -18.12 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.679008 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
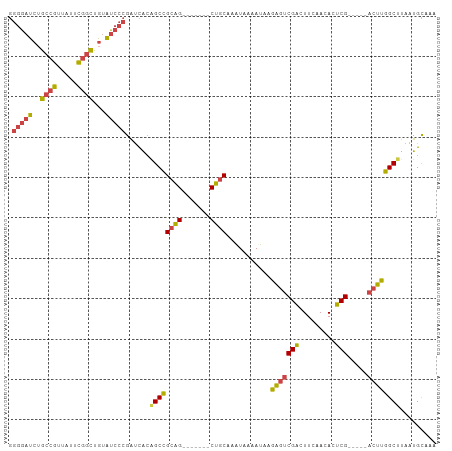
>dm3.chr3L 496969 94 - 24543557 GGGGAUCUGCCGUUAUUCGGCUGUAUCCCGAUCGCAGCCGCAG-------CUGCAAAUAAAAUAAGAGUCGACUUCAACACUCG-----ACUUGGCUUAAUGCAAA .((((((.((((.....)))).).)))))....(((((....)-------))))...........(((((((.........)))-----)))).((.....))... ( -32.00, z-score = -2.94, R) >droPer1.super_23 713965 105 + 1662726 -GGGAUCCUCCGUUAUGUGGAGAGGCAGAGGCUGAGGCUGAGGCAGAGGCCUACAGCUGCGAGAAAUUGCGGCUUCAACACUCGACACAACGUGGCUUAAUGUAAA -.((.....))....((((..((((..((((((..(((((((((....)))).)))))((((....))))))))))..).)))..))))((((......))))... ( -39.80, z-score = -2.31, R) >droSim1.chr2R 8715679 94 - 19596830 GGGGAUCUGCCGUUAUUCGGCUGUAUCCCGAUCACAGCCGCAG-------CUGCAAAUAAAAUAAGAGUCGACUUCAACACUCG-----ACUUGGCUUAAUGCAAA .((((((.((((.....)))).).)))))......((((((..-------..))...........(((((((.........)))-----))))))))......... ( -29.50, z-score = -2.68, R) >droSec1.super_2 518757 94 - 7591821 GGGGAUCUGCCGUUAUUCGGCUGUAUCCCGAUCACAGCCGCAG-------CUGCAAAUAAAAUAAGAGUCGACUUCAACACUCG-----ACUUGGCUUAAUGCAAA .((((((.((((.....)))).).)))))......((((((..-------..))...........(((((((.........)))-----))))))))......... ( -29.50, z-score = -2.68, R) >droYak2.chr3L 475482 94 - 24197627 GGGGAUCUGCCGUUAUUCCGCUGUAUCCCGAUCACCGCCGCAG-------CUGCAAAUAAAAUAAGAGUCGACUUCAACACUCG-----ACUUGGCUUAAUGCCAA .((((((.((.(.....).)).).))))).......(((((..-------..))...........(((((((.........)))-----))))))).......... ( -22.90, z-score = -0.78, R) >droEre2.scaffold_4784 494780 94 - 25762168 GGGGAUCUGCCGUUAUUCGCCUGUAUCCCGAUCACAGCCGCAG-------CUGCAAAUAAAAUAAGAGUCGACUUCAACACUCG-----ACUUGGCUUAAUGCAAA .((((((.(.((.....)).).).)))))......((((((..-------..))...........(((((((.........)))-----))))))))......... ( -22.40, z-score = -0.68, R) >consensus GGGGAUCUGCCGUUAUUCGGCUGUAUCCCGAUCACAGCCGCAG_______CUGCAAAUAAAAUAAGAGUCGACUUCAACACUCG_____ACUUGGCUUAAUGCAAA .(((((..((((.....))))...)))))......((((((((.......))))...........((((..........))))..........))))......... (-17.06 = -18.12 + 1.06)


| Location | 497,034 – 497,130 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 78.14 |
| Shannon entropy | 0.41760 |
| G+C content | 0.51409 |
| Mean single sequence MFE | -30.17 |
| Consensus MFE | -15.01 |
| Energy contribution | -18.40 |
| Covariance contribution | 3.39 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.885751 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 497034 96 + 24543557 GGGAUACAGCCGAAUAACGGCAGAUCCCCCCAUUAUGUUGAGCCACCCACUCAAGCCAGGUGGAGUGGAAAGGAUGAAACUCCAGAUC-AUAAAGGA-- (((((...((((.....))))..))))).((.(((((.....(((((((((.......))))).))))...(((......)))....)-)))).)).-- ( -33.60, z-score = -3.14, R) >droSim1.chr2R 8715744 96 + 19596830 GGGAUACAGCCGAAUAACGGCAGAUCCCCCCAUUAUGUUGAGCCACCCACUCAAGCCAGGUGGAGUGGAAAGGAUGAAACUCGAGAUC-AUAAAGGA-- (((((...((((.....))))..))))).((.(((((((((((((((((((.......))))).))))...........)))))...)-)))).)).-- ( -32.20, z-score = -2.80, R) >droSec1.super_2 518822 96 + 7591821 GGGAUACAGCCGAAUAACGGCAGAUCCCCCCAUUAUGUUGAGCCACCCACUCAAGCCAGGUGGAGUGGAAAGGAUGAAACUCGAGAUC-AUAAAGGA-- (((((...((((.....))))..))))).((.(((((((((((((((((((.......))))).))))...........)))))...)-)))).)).-- ( -32.20, z-score = -2.80, R) >droYak2.chr3L 475547 96 + 24197627 GGGAUACAGCGGAAUAACGGCAGAUCCCCCCAUUAUGUUGAGCCACCCACUCAAGCCAGGUGGAGUGGAAAGGAUGAAACUCGAGAUC-AUAAAGGA-- (((((...((.(.....).))..))))).((.(((((((((((((((((((.......))))).))))...........)))))...)-)))).)).-- ( -27.90, z-score = -1.16, R) >droEre2.scaffold_4784 494845 96 + 25762168 GGGAUACAGGCGAAUAACGGCAGAUCCCCCCAUUAUGUCGAGCCACCCACUCAAGCCAGGUGGAGUGGAAAGGAUGAAACUCGAGAUC-AUGAAGGA-- (((((...(.((.....)).)..))))).((.(((((((((((((((((((.......))))).))))...........)))))...)-)))).)).-- ( -29.80, z-score = -1.33, R) >droVir3.scaffold_13049 3294151 86 + 25233164 -----------GAAGCAUCUCGUAUAACGGCAUUUAAC--AGCCACCCACUCGGCCGGGGCAUGCCCCAUAAGGCAACUUGCCACAUUUAUGCACAAUA -----------..........((((((.(((.......--.)))........(((.((((....))))..(((....))))))....))))))...... ( -25.30, z-score = -2.03, R) >consensus GGGAUACAGCCGAAUAACGGCAGAUCCCCCCAUUAUGUUGAGCCACCCACUCAAGCCAGGUGGAGUGGAAAGGAUGAAACUCGAGAUC_AUAAAGGA__ (((((...((((.....))))..)))))....((((.((...(((((((((.......))))).))))..)).))))...................... (-15.01 = -18.40 + 3.39)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:53:08 2011