| Sequence ID | dm3.chr3L |
|---|---|
| Location | 493,666 – 493,744 |
| Length | 78 |
| Max. P | 0.860568 |

| Location | 493,666 – 493,744 |
|---|---|
| Length | 78 |
| Sequences | 10 |
| Columns | 82 |
| Reading direction | forward |
| Mean pairwise identity | 72.55 |
| Shannon entropy | 0.59791 |
| G+C content | 0.52441 |
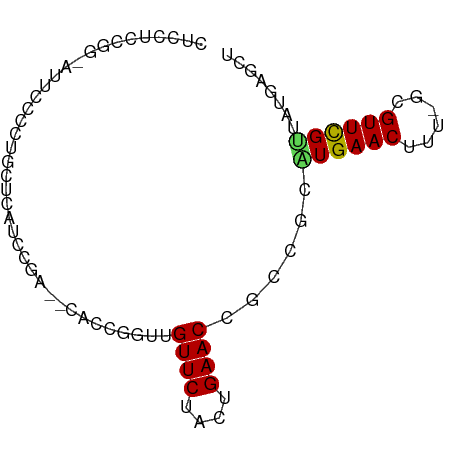
| Mean single sequence MFE | -19.46 |
| Consensus MFE | -6.39 |
| Energy contribution | -6.24 |
| Covariance contribution | -0.15 |
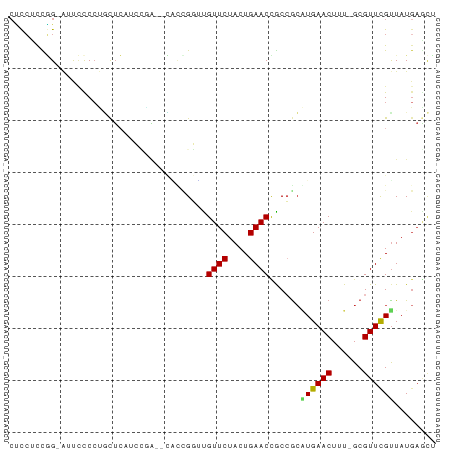
| Combinations/Pair | 1.30 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.860568 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
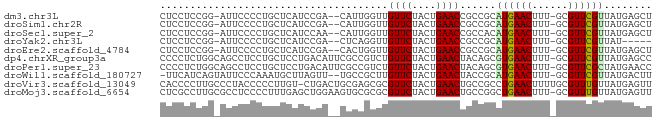
>dm3.chr3L 493666 78 + 24543557 CUCCUCCGG-AUUCCCCUGCUCAUCCGA--CAUUGGUUGUUCUACUGAACCGCCGCAUGAACUUU-GCGUUCGUUAUGAGCU .......((-.....)).((((((.(((--(...(((.((((....)))).)))(((.......)-))).)))..)))))). ( -21.60, z-score = -2.86, R) >droSim1.chr2R 8712415 78 + 19596830 CUCCUCCGG-AUUCCCCUGCUCAUCCGA--CAUUGGUUGUUCUACUGAACCGCCGCAUGAACUUU-GCGUUCGUUAUGAGCU .......((-.....)).((((((.(((--(...(((.((((....)))).)))(((.......)-))).)))..)))))). ( -21.60, z-score = -2.86, R) >droSec1.super_2 515500 78 + 7591821 CUCCUCCGG-AUUCCCCUGCUCAUCCAA--CAUUGGUUGUUCUACUGAACCGCCACAUGAACUUU-GCGUUCGUUAUGAGCU .......((-.....)).((((((..((--(..((((.((((....)))).))))...((((...-..))))))))))))). ( -21.60, z-score = -3.60, R) >droYak2.chr3L 472082 73 + 24197627 CUCCUCCGG-AUUCCCCUGCUCAUCCGA--CUCAGGUUGUUCUACUGAACCGCCGCAUGAACUUU-GCGUUCGUUAU----- ......(((-((..........))))).--....(((.((((....)))).)))..((((((...-..))))))...----- ( -16.70, z-score = -1.67, R) >droEre2.scaffold_4784 491523 78 + 25762168 CUCCUCCGG-AUUCCCCUGCUCAUCCGA--CACUGGUUGUUCUACUGAACCGCCGCAUGAACUUU-GCGUUCGUUAUGAGCU .......((-.....)).((((((.(((--.((.(((.((((....)))).)))(((.......)-)))))))..)))))). ( -22.00, z-score = -3.03, R) >dp4.chrXR_group3a 546238 81 - 1468910 CCCCUCUGGCAGCCUCCUGCUCCUGACAUUCGCCGUCUGUUCUACUGAACUACAGCGUGAACUUU-GCGUUCGUUAUGAGCC ..................((((.((((...(((.((..((((....)))).)).))).((((...-..)))))))).)))). ( -20.60, z-score = -1.83, R) >droPer1.super_23 709738 81 - 1662726 CCCCUCUGGCAGCCUCCUGCUCCUGACAUUCGCCGUCUGUUCUACUGAACUACAGCGUGAACUUU-GCGUUCGCUAUGAACC ....((.((.(((.....))))).))..(((((.((..((((....)))).)).))((((((...-..))))))...))).. ( -19.10, z-score = -1.96, R) >droWil1.scaffold_180727 2414188 78 - 2741493 -UUCAUCAGUAUUCCCAAAUGCUUAGUU--UGCCGCUUGUUCUACUGAACUACCGCAUGAACUUU-GCGUUCGUUAUGACUU -......(((.......(((((..((((--(((.(...((((....))))..).)))..))))..-))))).......))). ( -14.64, z-score = -1.60, R) >droVir3.scaffold_13049 3289890 81 + 25233164 CACCCCUUGCCCUACCCCCUUGU-CUGACUGCGAGCGCGUUCUACUGAACUGCCGCCUGAACUUUUGCGUUUGUUAUGAGUU ......................(-(((((.(((.(((.((((....))))))))))..((((......)))))))).))... ( -14.20, z-score = -1.48, R) >droMoj3.scaffold_6654 1871834 81 - 2564135 CUCGCCUUGCGCCUCCCCUUUGAGCUGGAAGUGCGCGCGUUCUACUGAACUGCCGGCUGAACUUU-GCGUUUGUUAUGAGUU ((((((..((((((..((........)).)).))))((((((....)))).)).))).((((...-..)))).....))).. ( -22.60, z-score = -0.67, R) >consensus CUCCUCCGG_AUUCCCCUGCUCAUCCGA__CACCGGUUGUUCUACUGAACCGCCGCAUGAACUUU_GCGUUCGUUAUGAGCU ......................................((((....))))......((((((......))))))........ ( -6.39 = -6.24 + -0.15)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:53:06 2011