| Sequence ID | dm3.chr3L |
|---|---|
| Location | 491,864 – 491,959 |
| Length | 95 |
| Max. P | 0.696433 |

| Location | 491,864 – 491,959 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 69.01 |
| Shannon entropy | 0.62789 |
| G+C content | 0.45816 |
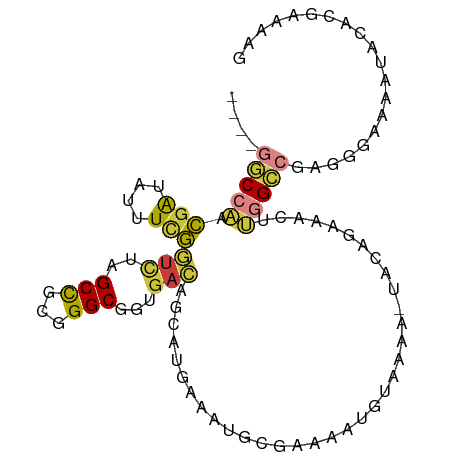
| Mean single sequence MFE | -21.09 |
| Consensus MFE | -10.48 |
| Energy contribution | -10.61 |
| Covariance contribution | 0.13 |
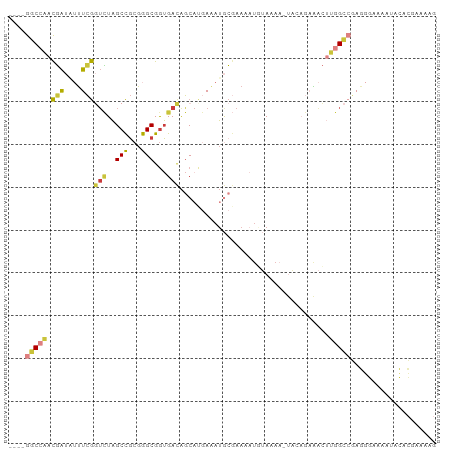
| Combinations/Pair | 1.57 |
| Mean z-score | -0.86 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.696433 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
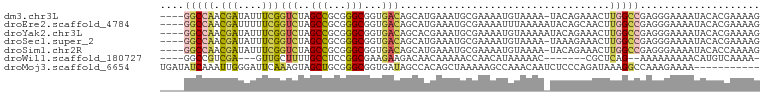
>dm3.chr3L 491864 95 + 24543557 ----GGCCAACGAUAUUUCGGUCUAGCCGCGGGCGGUGACAGCAUGAAAUGCGAAAAUGUAAAA-UACAGAAACUUGGCCGAGGGAAAAUACACGAAAAG ----((((((((.(((((((((...((((....))))....)).)))))))))....((((...-)))).....)))))).................... ( -23.40, z-score = -1.27, R) >droEre2.scaffold_4784 489789 96 + 25762168 ----GGCCAACGAUUUUUCGGUCUAGCCGCGGGCGGUGACAGCAUGAAAUGCGAAAAUUUAAAAAUACAGCAACUUGGCCGAGGGAAAAUACACGAAAAG ----(((((((((....)))(((..((((....))))))).((((...))))......................)))))).................... ( -22.20, z-score = -0.49, R) >droYak2.chr3L 470409 96 + 24197627 ----GGCCAACGAUAUUUCGGUCUAGCCGCGGGCGGUGACAGCACGAAAUGCGAAAAUGUAAAAAUACAGAAACUUGGCCGAGGGAAAAUACACGAAAAG ----((((((((.(((((((((...((((....))))....)).)))))))))....((((....)))).....)))))).................... ( -26.50, z-score = -2.23, R) >droSec1.super_2 513783 95 + 7591821 ----GGCCAACGAUAUUUCGGUCUAGCCGCGGGCGGUGACAGCAUGAAAUGCGAAAAUGUAAAA-UAAAGAAACUUGGCCGAGGGAAAAUACACGAAAAG ----(((((((((....)))(((..((((....))))))).((((...))))............-.........)))))).................... ( -22.20, z-score = -0.97, R) >droSim1.chr2R 8710674 95 + 19596830 ----GGCCAACGAUAUUUCGGUCUAGCCGCGGGCGGUGACAGCAUGAAAUGCGAAAAUGUAAAA-UACAGAAACUUGGCCGAGGGAAAAUACACCAAAAG ----((((((((.(((((((((...((((....))))....)).)))))))))....((((...-)))).....))))))..((.........))..... ( -24.60, z-score = -1.53, R) >droWil1.scaffold_180727 2411514 83 - 2741493 ----GGCCGUCGA---GUUGCUUUUGCCUCCGGCGAAGAAGACAACAAAAACCAACAUAAAAAC-------CGCUCAG--AAAAAAAAACAUGUCAAAA- ----.((((..((---(..((....)))))))))......((((....................-------.......--...........))))....- ( -10.46, z-score = 0.74, R) >droMoj3.scaffold_6654 1869473 89 - 2564135 UGAUAUCAAAUUGGGAUUCAAAGUAGCUGCGGGCGGUGAUAGCCACAGCUAAAAAGCCAAACAAUCUCCCAGAUAAAGGCCAAAGAAAA----------- ...((((....(((((.......((((((..(((.......))).))))))....(.....)....)))))))))..............----------- ( -18.30, z-score = -0.24, R) >consensus ____GGCCAACGAUAUUUCGGUCUAGCCGCGGGCGGUGACAGCAUGAAAUGCGAAAAUGUAAAA_UACAGAAACUUGGCCGAGGGAAAAUACACGAAAAG ....(((((.(((....)))(((..(((...)))...)))...................................))))).................... (-10.48 = -10.61 + 0.13)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:53:05 2011