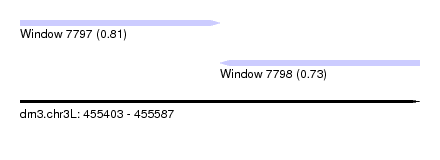
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 455,403 – 455,587 |
| Length | 184 |
| Max. P | 0.814422 |

| Location | 455,403 – 455,495 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 74.73 |
| Shannon entropy | 0.44986 |
| G+C content | 0.43026 |
| Mean single sequence MFE | -25.59 |
| Consensus MFE | -16.21 |
| Energy contribution | -14.83 |
| Covariance contribution | -1.38 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.814422 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
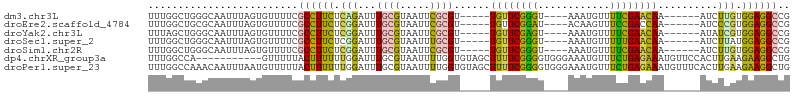
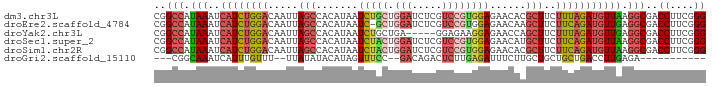
>dm3.chr3L 455403 92 + 24543557 UUUGGCUGGGCAAUUUAGUGUUUUCGCCUUCUCAGAUUUGCGUAAUUCGCGU-----UGUUCGGGU----AAAUGUUUUCGAACAA------AUCUUGUGGAGGCCG ....(((((.....)))))......((((((.((((((((((.....)))..-----.(((((((.----.......)))))))))------))).)).)))))).. ( -26.20, z-score = -1.70, R) >droEre2.scaffold_4784 453364 92 + 25762168 UUUGGCUGCGCAAUUUAGUGUUUUCGCCUUCUCGGAUUUGCGUAAUUCGCGU-----UGUUCGGAU----ACAAGUUUCCGACCAA------AUCCCGUGGAGGCCG ...((..((((......))))..))((((((.(((....(((.....))).(-----((.(((((.----.......))))).)))------...))).)))))).. ( -31.40, z-score = -2.51, R) >droYak2.chr3L 431786 92 + 24197627 UUUAGCUGGGCAAUUUAGUGUUUUCGCCUUCUCGGAUUUGCGUAAUUCGCGU-----UGUUCGAGU----AAAUGUUUUCGAACAA------AUAUCGUGGAGGCCG ....(((((.....)))))......((((((.(((....(((.....))).(-----((((((((.----.......)))))))))------...))).)))))).. ( -27.20, z-score = -2.06, R) >droSec1.super_2 477447 92 + 7591821 UUUGGCUGGGCAAUUUAGUGUUUUCGCCUUCUCGGAUUUGCGUAAUUUGCGU-----UGUUCGGGU----AAAUGUUUUUGAACAA------AUCUUAUGGAGGCCG ...((((..((((.((((((..((((......))))..))).))).)))).(-----((((((((.----.......)))))))))------..........)))). ( -22.60, z-score = -0.73, R) >droSim1.chr2R 8673907 92 + 19596830 UUUGGCUGGGCAAUUUAGUGUUUUCGCCUUCUCGGAUUUGCGUAAUUCGCGU-----UGUUCGGGU----AAAUGUUUUCGAACAA------AUCUUGUGGAGGCCG ....(((((.....)))))......((((((.((((((((((.....)))..-----.(((((((.----.......)))))))))------))).)).)))))).. ( -25.50, z-score = -1.21, R) >dp4.chrXR_group3a 496292 96 - 1468910 UUUGGCCA-----------GUUUUUACUUUUUUGGAUUUGCGUAAUUUUGGUGUAGCUUUUCGGGGUGGGAAAUGUUUCUGAGAAAUGUUCCACUUGAAGAAGGCUG ....((((-----------(...((((..............))))..))))).((((((((..((((((((...(((((...))))).))))))))...)))))))) ( -23.04, z-score = -0.77, R) >droPer1.super_23 660266 107 - 1662726 UUUGGCCAAACAAUUUAAUGUUUUUACUUUUUUGGAUUUGCGUAAUUUUGGUGUAGCUUUUCGGGGUGGGAAAUGUUUCUGAGAAAUGUUUCACUUGAAGAAGGCUG ...((((...(((..(.((((.....(......).....)))).)..))).......((((((((....((((((((((...)))))))))).)))))))).)))). ( -23.20, z-score = -0.80, R) >consensus UUUGGCUGGGCAAUUUAGUGUUUUCGCCUUCUCGGAUUUGCGUAAUUCGCGU_____UGUUCGGGU____AAAUGUUUUCGAACAA______AUCUUGUGGAGGCCG .........................((((((.(((...((((.....))))......((((((((............))))))))..........))).)))))).. (-16.21 = -14.83 + -1.38)
| Location | 455,495 – 455,587 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 75.22 |
| Shannon entropy | 0.48779 |
| G+C content | 0.47278 |
| Mean single sequence MFE | -22.76 |
| Consensus MFE | -15.14 |
| Energy contribution | -16.32 |
| Covariance contribution | 1.18 |
| Combinations/Pair | 1.33 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.734670 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
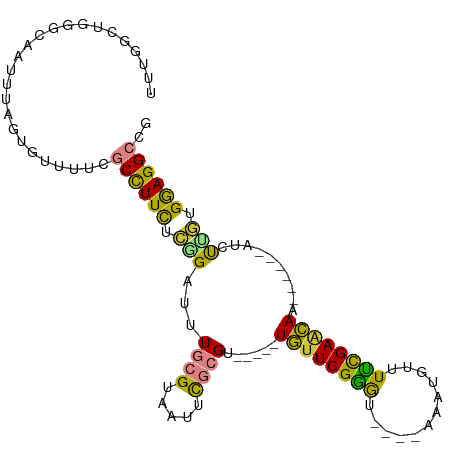
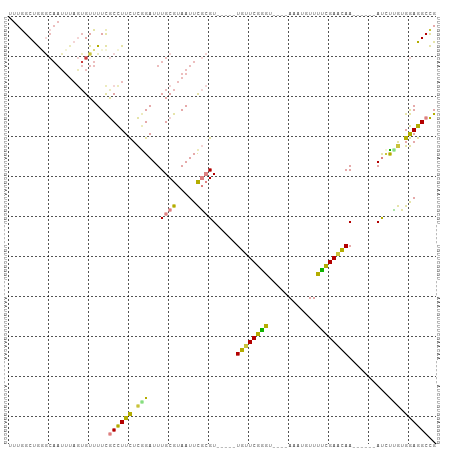
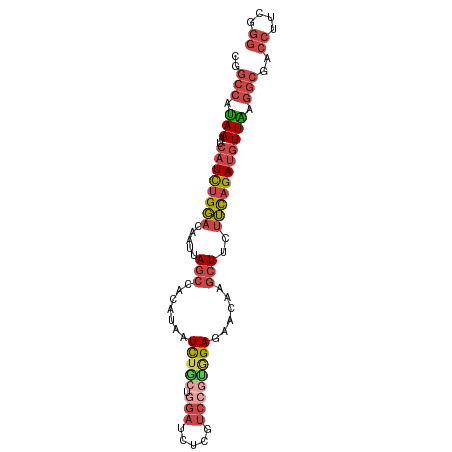
>dm3.chr3L 455495 92 - 24543557 CGGCCAUAAAUCAUCUGGACAAUUAGCCACAUAAUCUGCUGGAUCUCGUCCGUGGAGAACACGCUUCUUUAGAUGUUAAGGCGACCUUCGGG ..(((.(((..((((((((.....(((.......((..(.((((...)))))..))......)))..))))))))))).)))..((....)) ( -24.22, z-score = -0.92, R) >droEre2.scaffold_4784 453456 91 - 25762168 CGGCCAUAAAUCAUCUGGACAAUUAGCCACAUAAUC-GCUGGAUCUCGUCCGUGGAGAACAAGCUUCUUCAGAUGUUGAGGCGACCUUCGGG ..(((.(((..(((((((((......((((......-).))).....))))..((((((.....)))))))))))))).)))..((....)) ( -24.50, z-score = -0.48, R) >droYak2.chr3L 431878 87 - 24197627 CGGCCAUAAAUCAUCUGGACAAUUAGCCACAUAAUCUGCUGA-----GGAGAAGGAGAACCAGCUUCUUUAGAUGUUGAGGCGACCUUCGGG ..(((.(((..((((((((...(((((..........)))))-----((((..((....))..))))))))))))))).)))..((....)) ( -22.30, z-score = -0.52, R) >droSec1.super_2 477539 92 - 7591821 CGGCCAUAAAUCAUCUGGACAAUUAGCCACAUAAUCUACUGGAUCUCGUCCGUGGAGAACAUGCUUCUUCAGAUGUUAAGGCGACCUUCGGG ..(((.(((..((((((((.....(((.......(((((.((((...)))))))))......)))..))))))))))).)))..((....)) ( -27.22, z-score = -2.15, R) >droSim1.chr2R 8673999 92 - 19596830 CGGCCAUAAAUCAUCUGGACAAUUAGCCACAUAAUCUACUGGAUCUCGUCCGUGGAGAACACGCUUCUUCAGAUGUUAAGGCGACCUUCGGG ..(((.(((..((((((((.....(((.......(((((.((((...)))))))))......)))..))))))))))).)))..((....)) ( -27.22, z-score = -2.25, R) >droGri2.scaffold_15110 4149516 74 - 24565398 ---CGGCAAAUCAUUUGUUU--UUAUAUACAUAGUUUCC--GACAGACUCUUGAGAUUUCUUGCUGCUGCUGACCUUGAGA----------- ---(((.((((....(((..--......)))..))))))--)......(((..((...((..((....)).)).))..)))----------- ( -11.10, z-score = 0.36, R) >consensus CGGCCAUAAAUCAUCUGGACAAUUAGCCACAUAAUCUGCUGGAUCUCGUCCGUGGAGAACAAGCUUCUUCAGAUGUUAAGGCGACCUUCGGG ..(((.(((..((((((((.....(((.......(((((.(((.....))))))))......)))..))))))))))).)))..((....)) (-15.14 = -16.32 + 1.18)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:52:59 2011