| Sequence ID | dm3.chr3L |
|---|---|
| Location | 449,816 – 449,948 |
| Length | 132 |
| Max. P | 0.859114 |

| Location | 449,816 – 449,948 |
|---|---|
| Length | 132 |
| Sequences | 6 |
| Columns | 141 |
| Reading direction | reverse |
| Mean pairwise identity | 66.35 |
| Shannon entropy | 0.63170 |
| G+C content | 0.38053 |
| Mean single sequence MFE | -29.88 |
| Consensus MFE | -8.99 |
| Energy contribution | -11.72 |
| Covariance contribution | 2.73 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.859114 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
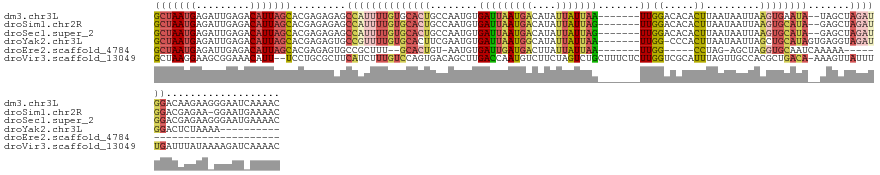
>dm3.chr3L 449816 132 - 24543557 GCUAAUGAGAUUGAGACAUUAGCACGAGAGAGCCAUUUUGUGCACUGCCAAUGUGAUUAAUGACAUAUUAUUAA-------UUGGACACACUUAAUAAUUAAGUGAAUA--UAGCUAGAUGGACAAGAAGGGAAUCAAAAC (((((((.........)))))))....((...((.((((((.((((((...((((((((((((....)))))))-------))..)))((((((.....))))))....--..)).)).)).)))))).))...))..... ( -30.10, z-score = -3.24, R) >droSim1.chr2R 8668356 131 - 19596830 GCUAAUGAGAUUGAGACAUUAGCACGAGAGAGCCAUUUUGUGCACUGCCAAUGUGAUUAAUGACAUAUUAUUAG-------UUGGACACACUUAAUAAUUAAGUGCAUA--GAGCUAGAUGGACGAGAA-GGAAUGAAAAC (((((((.........))))))).(....)..((.((((((.(((((((.(((.(((((((((....)))))))-------)).....((((((.....))))))))).--).)).)).)).)))))).-))......... ( -28.90, z-score = -2.69, R) >droSec1.super_2 471952 132 - 7591821 GCUAAUGAGAUUGAGACAUUAGCACGAGAGAGCCAUUUUGUGCACUGCCAAUGUGAUUAAUGACAUAUUAUUAG-------UUGGACACACUUAAUAAUUAAGUGCAUA--GAGCUAGAUGGACGAGAAGGGAAUGAAAAC (((((((.........))))))).(....)..((.((((((.(((((((.(((.(((((((((....)))))))-------)).....((((((.....))))))))).--).)).)).)).)))))).)).......... ( -29.10, z-score = -2.56, R) >droYak2.chr3L 426097 123 - 24197627 GCUAAUGAGAUUGAGACAUUAGCACGAGAGUGCCGUUUUGUGCACUUCGAAUGUGAUUAAUGGCAUAUUAUUAA-------UUGG-CCCACUUAAUAAUUAGCUGCAUAGUGAGGUAGAUGGACUCUAAAA---------- (((((((.........)))))))...(((((((((((.....(((.......)))...)))))).........(-------((.(-((((((................)))).))).)))..)))))....---------- ( -29.69, z-score = -1.04, R) >droEre2.scaffold_4784 447819 99 - 25762168 GCUAAUGAGAUUGAGACAUUAGCACGAGAGUGCCGCUUU--GCACUGU-AAUGUGAUUGAUGACUUAUUAUUAA-------UUGG-----CCUAG-AGCUAGGUGCAAUCAAAAA-------------------------- ........(((((..((((((((((....)))).((...--))....)-)))))(((((((((....)))))))-------)).(-----((((.-...))))).))))).....-------------------------- ( -25.10, z-score = -1.59, R) >droVir3.scaffold_13049 3218494 138 - 25233164 GCUAAGGAAGCGGAAACAUU--UCCUGCGCUUCAUCUUUGUCCAGUGACAGCUUGACCAAUGUCUUCUAGUCUGCUUUCUCUUGGUCGCAUUUAGUUGCCACGCUGACA-AAAGUUAUUUUGAUUUAUAAAAGAUCAAAAC ((..((((((.(....).))--))))..))......((((((..(((.((((((((((((.(......((....))....)))))))).....))))).)))...))))-)).....(((((((((.....))))))))). ( -36.40, z-score = -2.04, R) >consensus GCUAAUGAGAUUGAGACAUUAGCACGAGAGAGCCAUUUUGUGCACUGCCAAUGUGAUUAAUGACAUAUUAUUAA_______UUGGACACACUUAAUAAUUAAGUGCAUA__GAGCUAGAUGGACGAGAA_GGAAU_AAAAC (((((((.........))))))).........((((((((((((((.((((.....(((((((....))))))).......))))................)))))))).......))))))................... ( -8.99 = -11.72 + 2.73)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:52:58 2011