| Sequence ID | dm3.chr3L |
|---|---|
| Location | 444,783 – 444,902 |
| Length | 119 |
| Max. P | 0.956425 |

| Location | 444,783 – 444,902 |
|---|---|
| Length | 119 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.07 |
| Shannon entropy | 0.50300 |
| G+C content | 0.54733 |
| Mean single sequence MFE | -43.11 |
| Consensus MFE | -24.96 |
| Energy contribution | -25.18 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.956425 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
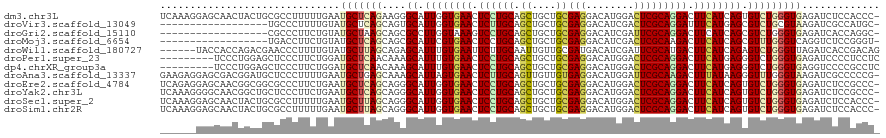
>dm3.chr3L 444783 119 + 24543557 UCAAAGGAGCAACUACUGCGCCUUUUUGAAUGCUCAGAAGGGCAUUGGUGAACUCCUGCAGCUGCUGCGAGGACAUGGACUCGCAGGACUUCAUCAGUGUCUGGGUGAGAUCUCCACCC- ...((((.(((.....))).))))(((((....))))).(((((((((((((.((((((.((....))(((........))))))))).)))))))))))))(((((.(....))))))- ( -50.50, z-score = -3.29, R) >droVir3.scaffold_13049 3212233 101 + 25233164 ------------------UGCCCUUUUGUAUGCUCAGCAGUGCAUUGGUGAACUCUUGCAGCUGCUGCGAGGACAUCGACUCGCAGGAUUUCAUGAGCGUCUGCGUAAGAUCGCCAUGC- ------------------...(((...........(((((((((..((....))..)))).)))))(((((........))))))))....((((.((((((.....))).))))))).- ( -35.40, z-score = -0.63, R) >droGri2.scaffold_15110 4135722 101 + 24565398 ------------------CGCCCUUCUGUAUGCUAAGCAGCGCCUUGGUAAAGUCCUGCAGCUGCUGCGAGGACAUCGAUUCGCAGGACUUCAUCAGCGUCUGGGUGAGAUCACCAGGC- ------------------.(((...............((((((..((((.(((((((((.((....))(((........)))))))))))).))))))).)))((((....)))).)))- ( -38.20, z-score = -1.17, R) >droMoj3.scaffold_6654 1804979 101 - 2564135 ------------------UGACCUUCUGUAUGCUCAGCAGCGCAUUCGUGAACUCCUGCAGCUGCUGCGAGGACAUCGACUCGCAAGACUUCAUCAGCGUUUGGGUCAGGUCUCCGGGU- ------------------.(((((((((.(((((.(((((((((...(....)...))).))))))(((((........)))))...........))))).))))..))))).......- ( -34.20, z-score = -0.21, R) >droWil1.scaffold_180727 2347226 114 - 2741493 ------UACCACCAGACGAACCCUUUUGUAUGCUUAGCAGAGCAUUUGUGAAUUCUUGCAAUUGUUGCGAUGACAUCGAUUCGCAUGACUUCAUCAGAGUCUGGGUUAGAUCACCGACAG ------.....((((((.............(((((....)))))((((((((.((.((((((((.((......)).))))).))).)).)))).))))))))))((..(....)..)).. ( -29.00, z-score = -1.21, R) >droPer1.super_23 649051 111 - 1662726 ---------UCCCUGGAGCUCCCUUCUGGAUGCUCAACAAAGCAUUUGUGAACUCCUGCAGCUGCUGCGAGGACAUGGACUCGCAGGACUUCAUGAGGGUCUGGGUGAGAUCCCCUCCUC ---------.....((((...(((((.(((((((......)))))))(((((.((((((.((....))(((........))))))))).))))))))))...(((......))))))).. ( -41.40, z-score = -1.00, R) >dp4.chrXR_group3a 485214 111 - 1468910 ---------UCCCUGGAGCUCCCUUCUGGAUGCUCAACAAAGCAUUUGUGAACUCCUGCAGCUGCUGCGAGGACAUGGACUCGCAGGACUUCAUGAGGGUCUGGGUGAGGUCCCCGCCUC ---------..((..((...(((((..(((((((......)))))))(((((.((((((.((....))(((........))))))))).))))))))))))..)).(((((....))))) ( -44.00, z-score = -1.21, R) >droAna3.scaffold_13337 23181029 119 - 23293914 GAAGAGGAGCGACGGAUGCUCCCUUUUGAAUGCUGAGCAAAGCAUUAGUGAACUCUUGCAGUUGUUGUGAGGACAUGGAUUCGCAAGACUUUAUAAGGGUUUGGGUAAGAUCGCCCCCG- ((((.((((((.....))))))))))..((((((......))))))...(((((((((.((((.(((((((........)))))))))))...)))))))))((((......))))...- ( -37.10, z-score = -1.47, R) >droEre2.scaffold_4784 440973 119 + 25762168 UCAGAGGAGCAACGGCGGCGCCCUUCUGAAUGCUCAGCAGGGCAUUGGUGAACUCCUGCAGCUGCUGCGAGGACAUGGACUCGCAGGACUUCAUCAGUGUCUGGGUGAGAUCUCCGCCC- .............((((((((((..((((....))))..(((((((((((((.((((((.((....))(((........))))))))).))))))))))))))))))......))))).- ( -53.30, z-score = -1.89, R) >droYak2.chr3L 421025 119 + 24197627 UCAAAGGGGCAACGGCUGCUCCCUUCUGAAUGCUCAGCAGGGCAUUGGUGAACUCCUGCAGCUGCUGCGAGGACAUGGACUCGCAGGACUUCAUCAGUGUCUGGGUGAGAUCUCCGCCC- (((((((((((.....))).))))).)))...((((.(.(((((((((((((.((((((.((....))(((........))))))))).))))))))))))).).))))..........- ( -53.80, z-score = -2.55, R) >droSec1.super_2 467030 119 + 7591821 UCAAAGGAGCAACUACUGCGCCUUUUUGAAUGCUUAGCAGGGCAUUGGUGAACUCCUGCAGCUGCUGCGAGGACAUGGACUCGCAGGACUUCAUCAGUGUCUGGGUGAGAUCUCCACCC- (((((((.(((.....))).)))..)))).(((...)))(((((((((((((.((((((.((....))(((........))))))))).)))))))))))))(((((.(....))))))- ( -50.20, z-score = -3.20, R) >droSim1.chr2R 8663412 119 + 19596830 UCAAAGGAGCAACUACUGCGCCUUUUUGAAUGCUUAGCAGGGCAUUGGUGAACUCCUGCAGCUGCUGCGAGGACAUGGACUCGCAGGACUUCAUCAGUGUCUGGGUGAGAUCUCCACCC- (((((((.(((.....))).)))..)))).(((...)))(((((((((((((.((((((.((....))(((........))))))))).)))))))))))))(((((.(....))))))- ( -50.20, z-score = -3.20, R) >consensus ______GAGCAACUGCUGCGCCCUUCUGAAUGCUCAGCAGGGCAUUGGUGAACUCCUGCAGCUGCUGCGAGGACAUGGACUCGCAGGACUUCAUCAGUGUCUGGGUGAGAUCUCCACCC_ ..............................(((((((....((.((((((((.((((((.((....))(((........))))))))).)))))))).)))))))))............. (-24.96 = -25.18 + 0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:52:56 2011