| Sequence ID | dm3.chr3L |
|---|---|
| Location | 437,806 – 437,898 |
| Length | 92 |
| Max. P | 0.975666 |

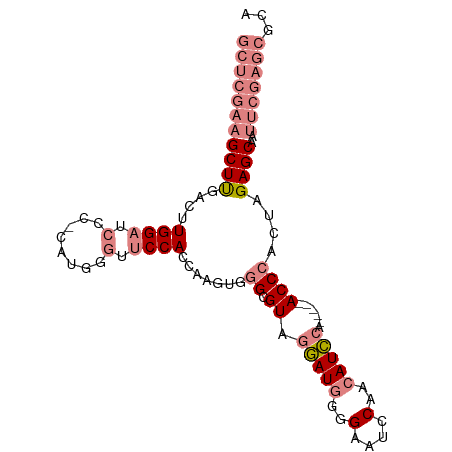
| Location | 437,806 – 437,898 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 75.36 |
| Shannon entropy | 0.44419 |
| G+C content | 0.58510 |
| Mean single sequence MFE | -36.81 |
| Consensus MFE | -15.71 |
| Energy contribution | -17.67 |
| Covariance contribution | 1.96 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.975666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 437806 92 + 24543557 GCUCGAAGCUUGACUUGGAUCCC-CAUGGGUUCCACCAAGUGGGCGUAGGAUGGGGAAUCCAACAUCCA-----ACCCACUAGAGCAAUUCGAGCGCA (((((((((((....((((.(((-...))).))))...((((((....(((((..(....)..))))).-----.)))))).))))..)))))))... ( -39.50, z-score = -3.65, R) >droEre2.scaffold_4784 434034 90 + 25762168 GCUCGAAGCUUGACUUGGAUCCC-C-UGUGUUCCAGCA-GUGGGCGUAGGAUGGGGAAUCCAACAUCCA-----ACCCACUGGAGCAAUUCGAGCGCA (((((((((((...(((((....-.-.....)))))((-(((((....(((((..(....)..))))).-----.)))))))))))..)))))))... ( -37.90, z-score = -2.76, R) >droYak2.chr3L 413949 95 + 24197627 GCUCGAAGCUCGACUUGGAUCCC-CAUGGGCUCCACCAAGUGGGCGUAGGAUGGGGAAUCCAACAUCCACCA--ACCCACCAGAGCAAUUCGAGCGCA (((((((((((....((((.(((-...))).))))....(((((.((.(((((..(....)..)))))))..--.)))))..))))..)))))))... ( -41.10, z-score = -3.77, R) >droSec1.super_2 460232 92 + 7591821 GCUCGAAGCUUGACUUGGAUCCC-CAUGGGUUCCACCAAGUUGGCGUAGGAUGGGGAAUCCAACAUCCA-----ACCCACUAGAGCAAUUCGAGCGCA ((((((((((((((((((..(((-...))).....)))))))((.((.(((((..(....)..))))).-----))))....))))..)))))))... ( -36.90, z-score = -3.29, R) >droSim1.chr2R 8656421 92 + 19596830 GCUCGAAGCUUGACUUGGAUCCC-CAUGGGUUCCACCAAGUGGGCGUAGGAUGGGGAAUCCAACAUCCA-----ACCCACUAGAGCAAUUCGAGCGCA (((((((((((....((((.(((-...))).))))...((((((....(((((..(....)..))))).-----.)))))).))))..)))))))... ( -39.50, z-score = -3.65, R) >dp4.chrXR_group3a 477020 87 - 1468910 -------GCUCCGCUUGGUUGCAGCCCAAGCUCCAGC---CACGCGUUGCAUGCAGCCCCCACAAUGGAGUGCAACCCAC-AGAGCGAGUCGAGCGCA -------((((((((((((((.(((....))).))))---)..(.(((((((....((........)).))))))).)..-.)))))....))))... ( -31.40, z-score = -1.44, R) >droPer1.super_23 640941 87 - 1662726 -------GCUCCGCUUGGUUGCAGCCCAAGCUCCAGC---CACGCGUUGCAUGCAGCCCCCACAAUGGAGUGCAACCCAC-AGAGCGAGUCGAGCGCA -------((((((((((((((.(((....))).))))---)..(.(((((((....((........)).))))))).)..-.)))))....))))... ( -31.40, z-score = -1.44, R) >consensus GCUCGAAGCUUGACUUGGAUCCC_CAUGGGUUCCACCAAGUGGGCGUAGGAUGGGGAAUCCAACAUCCA_____ACCCACUAGAGCAAUUCGAGCGCA (((((((((((....(((...........(((..........)))...(((((..(....)..)))))........)))...))))..)))))))... (-15.71 = -17.67 + 1.96)

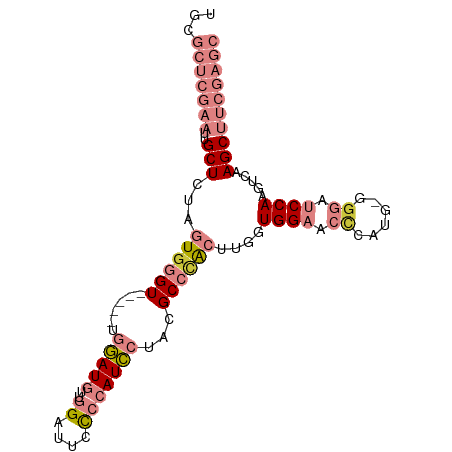
| Location | 437,806 – 437,898 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 75.36 |
| Shannon entropy | 0.44419 |
| G+C content | 0.58510 |
| Mean single sequence MFE | -36.87 |
| Consensus MFE | -18.18 |
| Energy contribution | -21.77 |
| Covariance contribution | 3.59 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.605366 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 437806 92 - 24543557 UGCGCUCGAAUUGCUCUAGUGGGU-----UGGAUGUUGGAUUCCCCAUCCUACGCCCACUUGGUGGAACCCAUG-GGGAUCCAAGUCAAGCUUCGAGC ...(((((((..((((.(((((((-----.(((((..((....)))))))...))))))).).((((.(((...-))).)))).....)))))))))) ( -37.90, z-score = -2.71, R) >droEre2.scaffold_4784 434034 90 - 25762168 UGCGCUCGAAUUGCUCCAGUGGGU-----UGGAUGUUGGAUUCCCCAUCCUACGCCCAC-UGCUGGAACACA-G-GGGAUCCAAGUCAAGCUUCGAGC ...(((((((..(((.((((((((-----.(((((..((....)))))))...))))))-)).((((.(...-.-..).)))).....)))))))))) ( -35.40, z-score = -2.12, R) >droYak2.chr3L 413949 95 - 24197627 UGCGCUCGAAUUGCUCUGGUGGGU--UGGUGGAUGUUGGAUUCCCCAUCCUACGCCCACUUGGUGGAGCCCAUG-GGGAUCCAAGUCGAGCUUCGAGC ...(((((((..((((.(((((((--....(((((..((....)))))))...)))))))...((((.(((...-))).))))....))))))))))) ( -41.40, z-score = -2.23, R) >droSec1.super_2 460232 92 - 7591821 UGCGCUCGAAUUGCUCUAGUGGGU-----UGGAUGUUGGAUUCCCCAUCCUACGCCAACUUGGUGGAACCCAUG-GGGAUCCAAGUCAAGCUUCGAGC ...(((((((..((((....))))-----..(((.((((((.((((((....((((.....))))......)))-))))))))))))....))))))) ( -35.90, z-score = -2.31, R) >droSim1.chr2R 8656421 92 - 19596830 UGCGCUCGAAUUGCUCUAGUGGGU-----UGGAUGUUGGAUUCCCCAUCCUACGCCCACUUGGUGGAACCCAUG-GGGAUCCAAGUCAAGCUUCGAGC ...(((((((..((((.(((((((-----.(((((..((....)))))))...))))))).).((((.(((...-))).)))).....)))))))))) ( -37.90, z-score = -2.71, R) >dp4.chrXR_group3a 477020 87 + 1468910 UGCGCUCGACUCGCUCU-GUGGGUUGCACUCCAUUGUGGGGGCUGCAUGCAACGCGUG---GCUGGAGCUUGGGCUGCAACCAAGCGGAGC------- ...(((((((((((...-)))))))(((((((......)))).)))......(((.((---(.((.(((....))).)).))).)))))))------- ( -34.80, z-score = 0.10, R) >droPer1.super_23 640941 87 + 1662726 UGCGCUCGACUCGCUCU-GUGGGUUGCACUCCAUUGUGGGGGCUGCAUGCAACGCGUG---GCUGGAGCUUGGGCUGCAACCAAGCGGAGC------- ...(((((((((((...-)))))))(((((((......)))).)))......(((.((---(.((.(((....))).)).))).)))))))------- ( -34.80, z-score = 0.10, R) >consensus UGCGCUCGAAUUGCUCUAGUGGGU_____UGGAUGUUGGAUUCCCCAUCCUACGCCCACUUGGUGGAACCCAUG_GGGAUCCAAGUCAAGCUUCGAGC ...(((((((..((((.(((((((......(((((..((....)))))))...))))))).).((((.(((....))).)))).....)))))))))) (-18.18 = -21.77 + 3.59)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:52:52 2011