| Sequence ID | dm3.chr3L |
|---|---|
| Location | 433,625 – 433,731 |
| Length | 106 |
| Max. P | 0.575250 |

| Location | 433,625 – 433,731 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.63 |
| Shannon entropy | 0.44985 |
| G+C content | 0.55119 |
| Mean single sequence MFE | -37.40 |
| Consensus MFE | -20.94 |
| Energy contribution | -23.12 |
| Covariance contribution | 2.17 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.575250 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
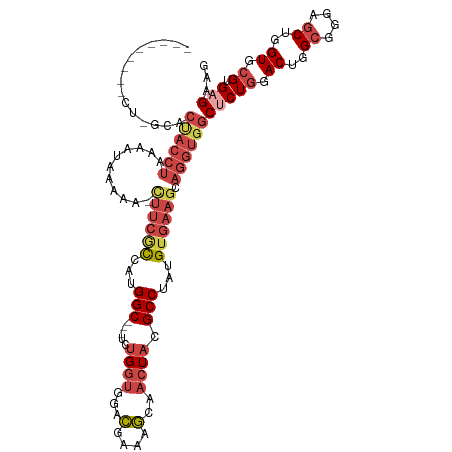
>dm3.chr3L 433625 106 - 24543557 --------------ACUACCUAAAAUAAACAGCUUCGUCAUGGCGGCUCUGGUGGACGAAAGCAACUACGCCUAUGUGAAGCAGGUGGCUCUGGACUGGCGGGAGCUGGUGCGCGAGAAG --------------.((((((..........((((((.(((((((....((((.(.(....)).)))))))).)))))))))))))))(((((.((..((....))..)).)).)))... ( -39.40, z-score = -2.01, R) >droSim1.chr2R 8652201 116 - 19596830 AUAGGUAUUCUCGCACUACCUAAAAUAACAA-CUUCGCCAUGGC---UCUGGUGGACGAAAGCAACUACGCCUAUGUGAAGCAGGUGGCCCUGGACUGGCGGGAGCUGGUGCGUGAGAAG .......(((((((.((((((..........-((((((...(((---..((((.(.(....)).)))).)))...)))))).))))))....(.((..((....))..)).)))))))). ( -41.00, z-score = -1.74, R) >droSec1.super_2 456014 116 - 7591821 AUAGGUAUUCUCGCACUACCUAAAAUAACAA-CUUCGCCAUGGC---UCUGGUGGACGAAAGCAACUACGCCUAUGUGAAGCAGGUGGCUCUGGACUGGCGGGAGCUGGUGCGUGAGAAG .......(((((((.((((((..........-((((((...(((---..((((.(.(....)).)))).)))...)))))).))))))....(.((..((....))..)).)))))))). ( -41.00, z-score = -2.03, R) >droYak2.chr3L 409645 116 - 24197627 GAAGGUAUUCUAGCACUACCUAAAAUAUAAA-CUUCACCAUGGC---UCUGGUGGACGAAAGCAACUACGCCUAUGUGAAGCAGGUGGCUCUGGACUGGCGGGAGCUGGUGCGCGAGAAG .......((((.((.((((((..........-((((((...(((---..((((.(.(....)).)))).)))...)))))).))))))....(.((..((....))..)).))).)))). ( -37.40, z-score = -1.47, R) >droEre2.scaffold_4784 429922 92 - 25762168 -------------------------CAGGAGUUUUCGCCAUGGCG---CUGGUGGACGAAGGCAACUACGCCUAUGUGAAGCAGGUGGCUCUGGACUGGCGGGAGCUGGUGCGGGAGAAG -------------------------......((((((((((....---(((.(..(((.((((......)))).)))..).)))))))).(((.((..((....))..)).)))))))). ( -35.10, z-score = -1.39, R) >droAna3.scaffold_13337 76007 103 - 23293914 ---------UCAGUUCCACCUUAAAAAGAA-----CAUCAUGGC---GUUGUCGGAGGAAGCGCAUUUCGCCUUUGCCAAGCAGGUGGCCCUCGACUGGCGGGAACUGGUCCGUGAGAAG ---------((((((((.((..........-----......))(---(((((((.((((((.((.....))))).((((......))))))))))).))))))))))))..(....)... ( -30.49, z-score = 0.54, R) >consensus _________CU_GCACUACCUAAAAUAAAAA_CUUCGCCAUGGC___UCUGGUGGACGAAAGCAACUACGCCUAUGUGAAGCAGGUGGCUCUGGACUGGCGGGAGCUGGUGCGUGAGAAG ...............((((((...........((((((...(((.....((((...(....)..)))).)))...)))))).))))))(((((.((..((....))..)).)).)))... (-20.94 = -23.12 + 2.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:52:51 2011