| Sequence ID | dm3.chr3L |
|---|---|
| Location | 333,571 – 333,669 |
| Length | 98 |
| Max. P | 0.542938 |

| Location | 333,571 – 333,669 |
|---|---|
| Length | 98 |
| Sequences | 9 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 71.48 |
| Shannon entropy | 0.54346 |
| G+C content | 0.55403 |
| Mean single sequence MFE | -32.04 |
| Consensus MFE | -18.10 |
| Energy contribution | -18.54 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.27 |
| Mean z-score | -0.84 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.542938 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 333571 98 + 24543557 CUGCUCGACGUGGUCUUGGUGGAGUACUGGCUCUGACGUUGGUUGCUGCCAUCGCGAACACUCGAGCAUCUGUGUUUGUGUGAAGUGGUGGAGUGGUU----------------- (..(((.((....((.(((((((((((..((......))..).)))).)))))).)).(((.((((((....)))))).))).....)).)))..)..----------------- ( -35.00, z-score = -1.39, R) >droSim1.chr2R 8559212 98 + 19596830 CUGCUCGACGUGGUCUUGGUGGAGUACUGGCUCUGACGUUGGUUGCUGCCAUCGCGAACACUCGAGCAUCUGUGUUCGUGUGAAGUGGUGGAGUGGUU----------------- (..(((.((....((.(((((((((((..((......))..).)))).)))))).)).(((.((((((....)))))).))).....)).)))..)..----------------- ( -37.10, z-score = -1.73, R) >droYak2.chr3L 313180 98 + 24197627 CUGCUCGACGUGGUCUUGGUGGAGUACUGGCUCUGACGUUGGUUGCUGCCAUCGCGAACACUCGAGCAUCUGUGGUCGUGUCAAGUGGUGGAGUGGUU----------------- (..(((.(((((.((.(((((((((((..((......))..).)))).)))))).)).))).(((.((....)).))).........)).)))..)..----------------- ( -33.70, z-score = -0.27, R) >droEre2.scaffold_4784 333502 98 + 25762168 CUGCUCGACGUGGUCUUGGUGGAAUAUUGGCUCUGACGUUGGUUGCUGCCAUCGCGAACACUCGAGCAUCUGUGGUCAUGUGAAGUGGUGAAGAGGUU----------------- .(((((((.(((.((.((((((...((..((......))..)).....)))))).)).))))))))))(((....((((.....))))...)))....----------------- ( -30.40, z-score = -0.32, R) >droAna3.scaffold_13337 281006 77 + 23293914 CUGCUCGACGUCGUCUUGGUGGAGUACUGGCUCUGACGCUGGCUGCUUCCCUCGCCCAUACUCGAGCACCUGGGGUC-------------------------------------- .(((((((.((.((...((.(((((((..((......))..).))))))))..))....))))))))).........-------------------------------------- ( -24.80, z-score = 0.31, R) >droSec1.super_1850 1519 98 + 4090 CUGCUCGACGUGGUCUUGGUGGAGUACUGGCUCUGACGUUGGUUGCUGCCAUCGCGAACACUCGAGCAUCUGUGUUCGUGUGAAGUGGUGGAGUGGUU----------------- (..(((.((....((.(((((((((((..((......))..).)))).)))))).)).(((.((((((....)))))).))).....)).)))..)..----------------- ( -37.10, z-score = -1.73, R) >droWil1.scaffold_180727 767628 79 - 2741493 CUGCUGGAUGUUGUUUUGGUGGAGUAUUGACUUUGCCGUUGAUUGCUACCAUCGCCCGAUCUCGAACAUCUAUUUGGUU------------------------------------ ....((((((((((..((((((...((..((......))..))..))))))..)).((....)))))))))).......------------------------------------ ( -21.60, z-score = -1.91, R) >droVir3.scaffold_13049 3041407 113 + 25233164 CUGCUGGACGUGGUCUUUGUGGAAUAUUGGCUCUGCCGCUGAUGGCUGCCCUCGCGCCCGCUCGAGCACCUACGG--GUGGGAAGGGGAAGGGUGAAACACACGGAAAACAGAUU .........((..(((.((((.....((.(((((((((....))))...((((...((((((((........)))--)))))..)))).))))).)).)))).)))..))..... ( -38.70, z-score = 0.03, R) >droMoj3.scaffold_6654 1658068 97 - 2564135 CUGCUGGAGGUGGUUUUGGUGGAGUAUUGACUCUGCCUCUGGUGGCUGCCAUCGCGCGAACUCGAGCAUCUAAAUGGACGGCAUGAGAAUCUGAUUA------------------ (..(..((((..(..(..(((...)))..)..)..))))..)..).((((...((.((....)).)).(((....))).))))..............------------------ ( -30.00, z-score = -0.53, R) >consensus CUGCUCGACGUGGUCUUGGUGGAGUACUGGCUCUGACGUUGGUUGCUGCCAUCGCGAACACUCGAGCAUCUGUGUUCGUGUGAAGUGGUGGAGUGGUU_________________ .((((.((.(((....(((((((((((..((......))..)).))).))))))....))))).))))............................................... (-18.10 = -18.54 + 0.45)


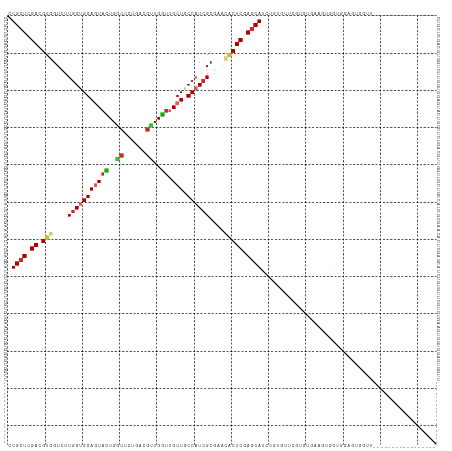
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:52:47 2011