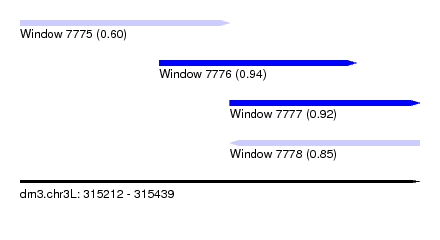
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 315,212 – 315,439 |
| Length | 227 |
| Max. P | 0.941170 |

| Location | 315,212 – 315,331 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 96.64 |
| Shannon entropy | 0.04630 |
| G+C content | 0.50140 |
| Mean single sequence MFE | -34.49 |
| Consensus MFE | -31.89 |
| Energy contribution | -32.22 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.596029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 315212 119 + 24543557 GUUUAGUGAAACAACCCGCAAGAUGAAAGCUCACAGCGAAGCCAUGCUGUUUAACGAGUAUUGUUUAAGUAAUGUGCUCGGGCUCCCCGAGCAGUUCUCCGGCCAUUCCGUUCAGUGAC ..(((.((((.......((..((.(((.((((((((((......)))))).....))))...............((((((((...)))))))).)))))..)).......)))).))). ( -31.34, z-score = -0.91, R) >droSim1.chr2R 8541040 119 + 19596830 GUUUAGUGAAACAGCCCGCAAGAUGAAAGCUCACAGCGAAGCCAUGCUGUUUUACGAGUAUUGUUUAAGUAAUGUGCUCGGGCUCCCCGAGCAGUUCUCCGGCCUUUCCGUUCACUGAC ..((((((((.......((..((.(((.((((((((((......)))))).....))))...............((((((((...)))))))).)))))..)).......)))))))). ( -35.24, z-score = -1.64, R) >droSec1.super_2 347003 119 + 7591821 GUUUAGUGAAACAGCCCGCAAGAUGAAAGCUCACAGCGAAGCCAUGCUGUUUAGCGAGUAUUGUUUAAGUAAUGUGCUCGGGCUCCCCGAGCAGUUCUACGGCCUUUCCGUUCACUGAC ..((((((((...(((.((((.((....(((.((((((......))))))..)))..)).))))....(((...((((((((...))))))))....)))))).......)))))))). ( -36.90, z-score = -1.77, R) >consensus GUUUAGUGAAACAGCCCGCAAGAUGAAAGCUCACAGCGAAGCCAUGCUGUUUAACGAGUAUUGUUUAAGUAAUGUGCUCGGGCUCCCCGAGCAGUUCUCCGGCCUUUCCGUUCACUGAC ..((((((((.....(((..(((.....((((((((((......)))))).....))))...............((((((((...))))))))..))).)))........)))))))). (-31.89 = -32.22 + 0.33)



| Location | 315,291 – 315,403 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 77.38 |
| Shannon entropy | 0.36614 |
| G+C content | 0.59537 |
| Mean single sequence MFE | -37.63 |
| Consensus MFE | -24.06 |
| Energy contribution | -25.00 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.941170 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 315291 112 + 24543557 GGGCUCCCCGAGCAGUUCUCCGGCCAUUCCGUUCAGUGACUCCAAUUCCGGCUGAAGCUGCAGCUGCACGCCAAACCCAAGCAACUGCAAGUCCAGCAAACCACUGAAUUCA .((((....(((.....))).)))).....((((((((....(......)((((..(((((((.(((.............))).)))).))).))))....))))))))... ( -29.32, z-score = -0.30, R) >droSim1.chr2R 8541119 112 + 19596830 GGGCUCCCCGAGCAGUUCUCCGGCCUUUCCGUUCACUGACUCCGAUUCCGGCUGCAGCUGCAGCUGCACGCCAAACCCAAGCAACUGCGAGUCCAGCAAACCACUGGAUUCA (((......(((((((....(((.....)))...)))).))).......(((((((((....)))))).)))...)))..((....))((((((((.......)))))))). ( -42.60, z-score = -3.15, R) >droSec1.super_2 347082 112 + 7591821 GGGCUCCCCGAGCAGUUCUACGGCCUUUCCGUUCACUGACUCCGAUUCCGGCUGCAGCUGCAGCUGCACACCAAACCCAAGCAACUGCGAGUCCAGCAAACCACUGGAUUCA (((......(((((((...((((.....))))..)))).))).......((.((((((....))))))..))...)))..((....))((((((((.......)))))))). ( -40.50, z-score = -3.13, R) >droYak2.chr3L 294607 88 + 24197627 GGGCUGCCCGAGCAGCUCUCCUGCCACUCCGUUCAGUGACUCCGAUUUCGGCUGCAGUUGCAGCUGCA-GCUGCAUGCCACUGGAUUCA----------------------- (((((((....)))))))............((((((((....((....))(((((((((((....)))-)))))).))))))))))...----------------------- ( -38.10, z-score = -2.05, R) >consensus GGGCUCCCCGAGCAGUUCUCCGGCCAUUCCGUUCACUGACUCCGAUUCCGGCUGCAGCUGCAGCUGCACGCCAAACCCAAGCAACUGCAAGUCCAGCAAACCACUGGAUUCA (((...)))..((((((((..((.......(((....))).........(((((((((....)))))).)))...))..)).))))))((((((((.......)))))))). (-24.06 = -25.00 + 0.94)


| Location | 315,331 – 315,439 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 80.09 |
| Shannon entropy | 0.32423 |
| G+C content | 0.55155 |
| Mean single sequence MFE | -38.06 |
| Consensus MFE | -22.74 |
| Energy contribution | -25.48 |
| Covariance contribution | 2.75 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.919185 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 315331 108 + 24543557 UCCAAUUCCGGCUGAAGCUGCAGCUGCACGCCAAACCCAAGCAACUGCAAGUCCAGCAAACCACUGAAUUCAGUUUCUUUCGAACAGCUGGGCUGCUCCAGCGGUUUG ..(((..(((.(((.((((((((.(((.............))).)))))((((((((.....((((....))))(((....)))..))))))))))).)))))).))) ( -37.92, z-score = -2.54, R) >droSim1.chr2R 8541159 108 + 19596830 UCCGAUUCCGGCUGCAGCUGCAGCUGCACGCCAAACCCAAGCAACUGCGAGUCCAGCAAACCACUGGAUUCAGUUUCUUGCGAACAGCUGUGCUGCUCCAGCGAUUUG ..(((.(((((..(((((.(((((((..(((.((((....((....))((((((((.......)))))))).))))...)))..)))))))))))).)).).)).))) ( -41.60, z-score = -2.92, R) >droSec1.super_2 347122 108 + 7591821 UCCGAUUCCGGCUGCAGCUGCAGCUGCACACCAAACCCAAGCAACUGCGAGUCCAGCAAACCACUGGAUUCAGUUUCUUGCACACAGCUGUGCUGCUCCAGCGAUUUG ..(((.(((((..(((((.(((((((..((..((((....((....))((((((((.......)))))))).))))..))....)))))))))))).)).).)).))) ( -37.00, z-score = -2.09, R) >droYak2.chr3L 294647 82 + 24197627 UCCGAUUUCGGCUGCAGUUGCAGCUGCA---------------GCUGCAUG---------CCACUGGAUUCAGUUUCUU--GAACAGCUGUGCGGCUGCAGUGGUUUG ........((((((((((....))))))---------------))))((.(---------((((((..(((((....))--)))(((((....)))))))))))).)) ( -35.70, z-score = -2.07, R) >consensus UCCGAUUCCGGCUGCAGCUGCAGCUGCACGCCAAACCCAAGCAACUGCAAGUCCAGCAAACCACUGGAUUCAGUUUCUUGCGAACAGCUGUGCUGCUCCAGCGAUUUG ..........((((.(((.(((((((...........((((.(((((..(((((((.......)))))))))))).))))....)))))))...))).))))...... (-22.74 = -25.48 + 2.75)


| Location | 315,331 – 315,439 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 80.09 |
| Shannon entropy | 0.32423 |
| G+C content | 0.55155 |
| Mean single sequence MFE | -41.85 |
| Consensus MFE | -29.52 |
| Energy contribution | -30.90 |
| Covariance contribution | 1.37 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.851129 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 315331 108 - 24543557 CAAACCGCUGGAGCAGCCCAGCUGUUCGAAAGAAACUGAAUUCAGUGGUUUGCUGGACUUGCAGUUGCUUGGGUUUGGCGUGCAGCUGCAGCUUCAGCCGGAAUUGGA ....((((((((((((.(((((..(((....)))((((....)))).....))))).))(((((((((.((.......)).)))))))))))))))).)))....... ( -47.20, z-score = -2.69, R) >droSim1.chr2R 8541159 108 - 19596830 CAAAUCGCUGGAGCAGCACAGCUGUUCGCAAGAAACUGAAUCCAGUGGUUUGCUGGACUCGCAGUUGCUUGGGUUUGGCGUGCAGCUGCAGCUGCAGCCGGAAUCGGA .((((((((((((((((...)))))((....)).......)))))))))))((..(((((((....))..)))))..)).((((((....)))))).(((....))). ( -46.60, z-score = -1.82, R) >droSec1.super_2 347122 108 - 7591821 CAAAUCGCUGGAGCAGCACAGCUGUGUGCAAGAAACUGAAUCCAGUGGUUUGCUGGACUCGCAGUUGCUUGGGUUUGGUGUGCAGCUGCAGCUGCAGCCGGAAUCGGA .......((((.(((((.((((((((..(...(((((((.((((((.....)))))).))((....))...))))).)..))))))))..)))))..))))....... ( -43.20, z-score = -0.88, R) >droYak2.chr3L 294647 82 - 24197627 CAAACCACUGCAGCCGCACAGCUGUUC--AAGAAACUGAAUCCAGUGG---------CAUGCAGC---------------UGCAGCUGCAACUGCAGCCGAAAUCGGA .......((((((..((...(((((((--(......))))..)))).)---------).((((((---------------....)))))).))))))(((....))). ( -30.40, z-score = -1.90, R) >consensus CAAACCGCUGGAGCAGCACAGCUGUUCGCAAGAAACUGAAUCCAGUGGUUUGCUGGACUCGCAGUUGCUUGGGUUUGGCGUGCAGCUGCAGCUGCAGCCGGAAUCGGA (((((((((((((((((...)))))((....)).......))))))))))))........((((((((.............))))))))........(((....))). (-29.52 = -30.90 + 1.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:52:42 2011