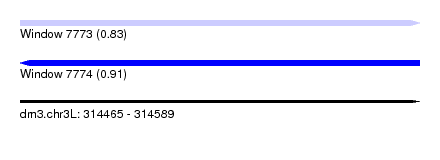
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 314,465 – 314,589 |
| Length | 124 |
| Max. P | 0.907940 |

| Location | 314,465 – 314,589 |
|---|---|
| Length | 124 |
| Sequences | 3 |
| Columns | 133 |
| Reading direction | forward |
| Mean pairwise identity | 64.91 |
| Shannon entropy | 0.48331 |
| G+C content | 0.35688 |
| Mean single sequence MFE | -29.83 |
| Consensus MFE | -15.80 |
| Energy contribution | -14.71 |
| Covariance contribution | -1.09 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.832356 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
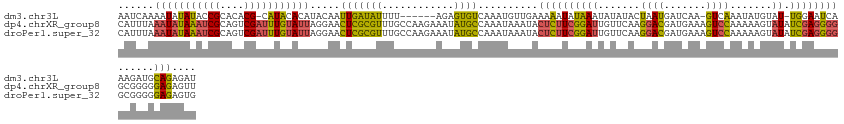
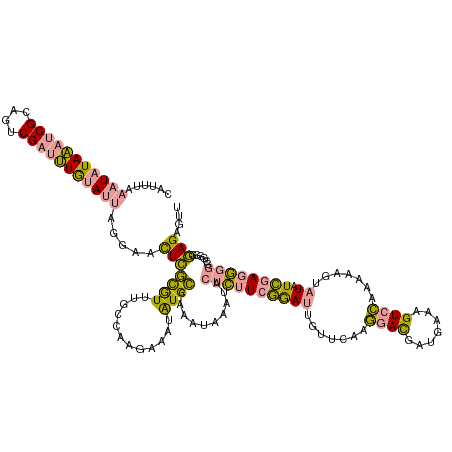
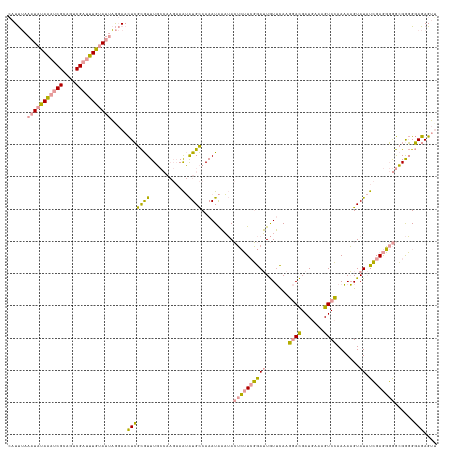
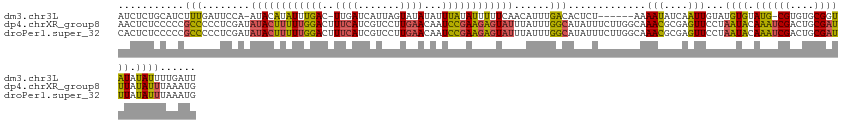
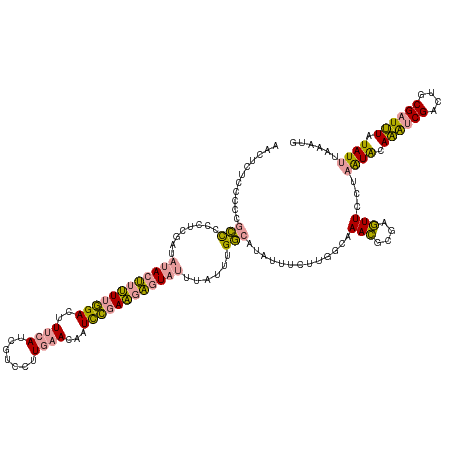
>dm3.chr3L 314465 124 + 24543557 AAUCAAAAUAUAUACCGCACACG-CAUACACAUACAAUUGAUAUUUU------AGAGUGUCAAAUGUUGAAAAAUAUAAAUAUAUACUAAUGAUCAA-GUCAAAUAUGUAU-UGGAAUCAAAGAUGCAGAGAU ......................(-(((...((.(((.((((((((..------..)))))))).))))).........((((((((....((((...-))))..)))))))-)..........))))...... ( -16.60, z-score = 0.21, R) >dp4.chrXR_group8 7802537 133 - 9212921 CAUUUAAAUAUAAAUCGCAGUCGAUUUGUAUUAGGAACUCGCGUUUGCCAAGAAAUAUGCCAAAUAAAUACUCUUCGGAUUGUUCAAGGACGAUGAAAGUCCAAAAAGUAUAUCGAGGGGGCGGGGGAGAGUU ......(((((((((((....)))))))))))...(((((.(.((((((.....((((((......((((.((....)).))))...((((.......)))).....))))))......)))))).).))))) ( -36.40, z-score = -2.55, R) >droPer1.super_32 65814 133 + 992631 CAUUUAAAUAUAAAUCGCAGUCGAUUUGUAUUAGGAACUCGCGUUUGCCAAGAAAUAUGCCAAAUAAAUACUCUUCGGAUUGUUCAAGGACGAUGAAAGUCCAAAAAGUAUAUCGAGGGGGCGGGGGAGAGUG ..((..(((((((((((....)))))))))))..))((((.(.((((((.....((((((......((((.((....)).))))...((((.......)))).....))))))......)))))).).)))). ( -36.50, z-score = -2.56, R) >consensus CAUUUAAAUAUAAAUCGCAGUCGAUUUGUAUUAGGAACUCGCGUUUGCCAAGAAAUAUGCCAAAUAAAUACUCUUCGGAUUGUUCAAGGACGAUGAAAGUCCAAAAAGUAUAUCGAGGGGGCGGGGGAGAGUU ......(((((((((((....))))))))))).....(((((((.(........).))))..........((((((((.........((((.......))))..........))))))))......))).... (-15.80 = -14.71 + -1.09)
| Location | 314,465 – 314,589 |
|---|---|
| Length | 124 |
| Sequences | 3 |
| Columns | 133 |
| Reading direction | reverse |
| Mean pairwise identity | 64.91 |
| Shannon entropy | 0.48331 |
| G+C content | 0.35688 |
| Mean single sequence MFE | -24.89 |
| Consensus MFE | -14.50 |
| Energy contribution | -14.07 |
| Covariance contribution | -0.43 |
| Combinations/Pair | 1.34 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.907940 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 314465 124 - 24543557 AUCUCUGCAUCUUUGAUUCCA-AUACAUAUUUGAC-UUGAUCAUUAGUAUAUAUUUAUAUUUUUCAACAUUUGACACUCU------AAAAUAUCAAUUGUAUGUGUAUG-CGUGUGCGGUAUAUAUUUUGAUU ....((((((...((......-((((((((.....-.(((.....((((((....))))))..)))(((.((((......------......)))).))))))))))).-)).)))))).............. ( -16.20, z-score = 1.14, R) >dp4.chrXR_group8 7802537 133 + 9212921 AACUCUCCCCCGCCCCCUCGAUAUACUUUUUGGACUUUCAUCGUCCUUGAACAAUCCGAAGAGUAUUUAUUUGGCAUAUUUCUUGGCAAACGCGAGUUCCUAAUACAAAUCGACUGCGAUUUAUAUUUAAAUG (((((......(((........((((((((((((..((((.......))))...))))))))))))......)))..........((....)))))))...((((.((((((....)))))).))))...... ( -29.54, z-score = -3.11, R) >droPer1.super_32 65814 133 - 992631 CACUCUCCCCCGCCCCCUCGAUAUACUUUUUGGACUUUCAUCGUCCUUGAACAAUCCGAAGAGUAUUUAUUUGGCAUAUUUCUUGGCAAACGCGAGUUCCUAAUACAAAUCGACUGCGAUUUAUAUUUAAAUG .((((......(((........((((((((((((..((((.......))))...))))))))))))......)))..........((....))))))....((((.((((((....)))))).))))...... ( -28.94, z-score = -2.88, R) >consensus AACUCUCCCCCGCCCCCUCGAUAUACUUUUUGGACUUUCAUCGUCCUUGAACAAUCCGAAGAGUAUUUAUUUGGCAUAUUUCUUGGCAAACGCGAGUUCCUAAUACAAAUCGACUGCGAUUUAUAUUUAAAUG ...........(((........((((((((((((..((((.......))))...))))))))))))......))).............(((....)))...((((.((((((....)))))).))))...... (-14.50 = -14.07 + -0.43)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:52:39 2011