
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 312,086 – 312,221 |
| Length | 135 |
| Max. P | 0.848597 |

| Location | 312,086 – 312,189 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.59 |
| Shannon entropy | 0.26579 |
| G+C content | 0.48911 |
| Mean single sequence MFE | -37.58 |
| Consensus MFE | -26.57 |
| Energy contribution | -27.01 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.848597 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 312086 103 + 24543557 AACUGCCAAUGGGUCAUCCCGUAUAGC-----------------UGCUGCUGAAAAGCGGAACACUCCCUUUAUGUGGCCAUCCCGGAAAUAAAAUUUGAAUACGGGUCAGCCAACGUGA ....((..(((((....)))))...))-----------------..(((((....)))))..........(((((((((...((((.................))))...))).)))))) ( -29.53, z-score = -1.13, R) >droYak2.chr3L 291350 119 + 24197627 AACUGCCAACGGGUCGUUCCGUAUGAAAACCGAAAGG-CUUUUCCGCUUCUGAAAAGCGGAGCACUCCCAUUGUGCGGCCAUCCCGGAAAUGAUAUUUGAAUACGGGACAGCCCAUGGGA .....(((..((((.(((((((((..((((((...((-((..(((((((.....)))))))((((.......))))))))....)))........)))..))))))))).)))).))).. ( -45.50, z-score = -3.02, R) >droSec1.super_2 343745 120 + 7591821 AACUGUCUAUGGGUCAUCCCGUAUAGCAACCGAAAGGGCUUAUCUGCUGCUGAAAAGCGGAACACUCCCUUUAUGUGGCCAUCCCGGAAAUAAUAUUUGAAUACGGGACAGCCAACGUGA ...(((.((((((....))))))..)))....((((((........(((((....)))))......))))))(((((((..(((((.................)))))..))).)))).. ( -37.67, z-score = -2.12, R) >droSim1.chr2R 8537746 120 + 19596830 AACUGCCUAUGGGUCAUCCCGUAUAGCAACCGAAAGGGCUUAUCUGUUGCUGAAAAGCGGAACACUCCCUUUAUGUGGCCAUCCCGGAAAUAAUAUUUGAAUACGGGACAGCCAACGUGA ..((((.((((((....))))))(((((.((....))((......)))))))....))))..........(((((((((..(((((.................)))))..))).)))))) ( -37.63, z-score = -2.04, R) >consensus AACUGCCAAUGGGUCAUCCCGUAUAGCAACCGAAAGG_CUUAUCUGCUGCUGAAAAGCGGAACACUCCCUUUAUGUGGCCAUCCCGGAAAUAAUAUUUGAAUACGGGACAGCCAACGUGA ........(((((....))))).......((....)).....((((((.......)))))).........(((((((((..(((((.................)))))..))).)))))) (-26.57 = -27.01 + 0.44)

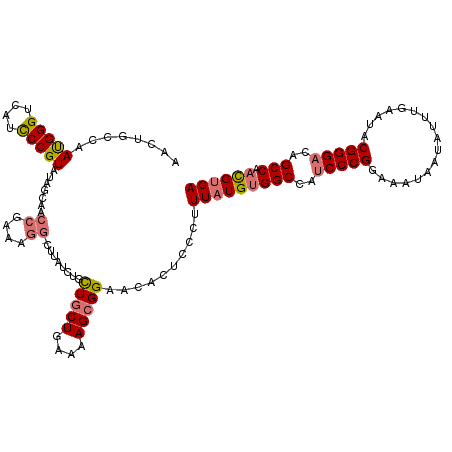

| Location | 312,086 – 312,189 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.59 |
| Shannon entropy | 0.26579 |
| G+C content | 0.48911 |
| Mean single sequence MFE | -35.91 |
| Consensus MFE | -30.19 |
| Energy contribution | -30.76 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.650454 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 312086 103 - 24543557 UCACGUUGGCUGACCCGUAUUCAAAUUUUAUUUCCGGGAUGGCCACAUAAAGGGAGUGUUCCGCUUUUCAGCAGCA-----------------GCUAUACGGGAUGACCCAUUGGCAGUU ....((.(((((.((((.................)))).))))))).....(((..(((((((......(((....-----------------)))...))))))).))).......... ( -25.33, z-score = 0.57, R) >droYak2.chr3L 291350 119 - 24197627 UCCCAUGGGCUGUCCCGUAUUCAAAUAUCAUUUCCGGGAUGGCCGCACAAUGGGAGUGCUCCGCUUUUCAGAAGCGGAAAAG-CCUUUCGGUUUUCAUACGGAACGACCCGUUGGCAGUU .......((((((((((.................))))))))))((.(((((((.....((((((((...))))))))((((-((....))))))............))))))))).... ( -45.13, z-score = -2.62, R) >droSec1.super_2 343745 120 - 7591821 UCACGUUGGCUGUCCCGUAUUCAAAUAUUAUUUCCGGGAUGGCCACAUAAAGGGAGUGUUCCGCUUUUCAGCAGCAGAUAAGCCCUUUCGGUUGCUAUACGGGAUGACCCAUAGACAGUU ....(((((((((((((.................)))))))))).......(((..(((((((......((((((.((.........)).))))))...))))))).)))...))).... ( -36.83, z-score = -1.66, R) >droSim1.chr2R 8537746 120 - 19596830 UCACGUUGGCUGUCCCGUAUUCAAAUAUUAUUUCCGGGAUGGCCACAUAAAGGGAGUGUUCCGCUUUUCAGCAACAGAUAAGCCCUUUCGGUUGCUAUACGGGAUGACCCAUAGGCAGUU ....(((((((((((((.................)))))))))).......(((..(((((((......((((((.((.........)).))))))...))))))).)))...))).... ( -36.33, z-score = -1.34, R) >consensus UCACGUUGGCUGUCCCGUAUUCAAAUAUUAUUUCCGGGAUGGCCACAUAAAGGGAGUGUUCCGCUUUUCAGCAGCAGAUAAG_CCUUUCGGUUGCUAUACGGGAUGACCCAUAGGCAGUU .......((((((((((.................)))))))))).......(((..(((((((......((((((.((.........)).))))))...))))))).))).......... (-30.19 = -30.76 + 0.56)

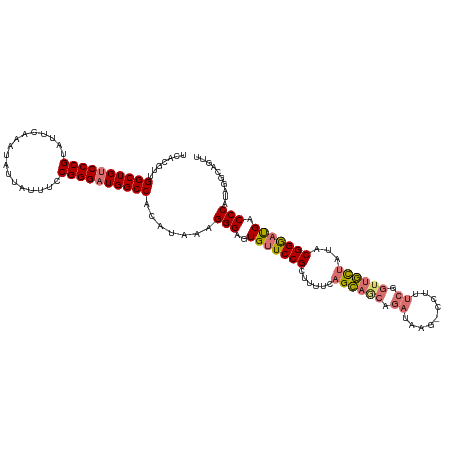
| Location | 312,112 – 312,221 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.33 |
| Shannon entropy | 0.28468 |
| G+C content | 0.50765 |
| Mean single sequence MFE | -37.66 |
| Consensus MFE | -25.25 |
| Energy contribution | -26.41 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.516503 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 312112 109 - 24543557 -----GCGCAAAUGAUGGUAGGACACCAGCCAGA---UGAUCACGUUGGCUGACCCGUAUUCAAAUUUUAUUUCCGGGAUGGCCACAUAAAGGGAGUGUUCCGCUUUUCAGCAGCAG--- -----..((...(((.((..((((((((((((.(---((....)))))))))...................((((...(((....)))...))))))))))..))..)))...))..--- ( -31.20, z-score = -0.85, R) >droEre2.scaffold_4784 304596 112 - 25762168 -----GCGCGGAUGUUGGUAGGACAGCAGCCAGA---AGAUCCCACUGGCUGUCCCGUAUGUAAAUAUCAUUUCCGGGAUGGCCACAUAGUGGGAGUGUUCCGCGUUUCGGGAGCGGAAG -----((((((((((((......)))))......---...(((((((((((((((((.(((.......)))...))))))))))....)))))))....)))))))((((....)))).. ( -48.90, z-score = -3.70, R) >droYak2.chr3L 291389 120 - 24197627 ACCGAGCGCGAAUGUUGGUAGGACACCAUGAUGACGUUGAUCCCAUGGGCUGUCCCGUAUUCAAAUAUCAUUUCCGGGAUGGCCGCACAAUGGGAGUGCUCCGCUUUUCAGAAGCGGAAA .((((((((.(((((((((.....)))).....)))))..((((((.((((((((((.................)))))))))).....)))))))))))).(((((...)))))))... ( -44.23, z-score = -1.88, R) >droSec1.super_2 343785 112 - 7591821 -----GCGCAAAUGAUAGUAGCACGCCAGCCAGA---UGAUCACGUUGGCUGUCCCGUAUUCAAAUAUUAUUUCCGGGAUGGCCACAUAAAGGGAGUGUUCCGCUUUUCAGCAGCAGAUA -----..((...(((.((((((((.((.....((---((....))))((((((((((.................))))))))))........)).)))))..)))..)))...))..... ( -31.33, z-score = -0.84, R) >droSim1.chr2R 8537786 112 - 19596830 -----GCGCAAAUGAUGGUAGGACACCAGCCAGA---UGAUCACGUUGGCUGUCCCGUAUUCAAAUAUUAUUUCCGGGAUGGCCACAUAAAGGGAGUGUUCCGCUUUUCAGCAACAGAUA -----..((...(((.((..((((((...((.((---((....))))((((((((((.................))))))))))........)).))))))..))..)))))........ ( -32.63, z-score = -1.21, R) >consensus _____GCGCAAAUGAUGGUAGGACACCAGCCAGA___UGAUCACGUUGGCUGUCCCGUAUUCAAAUAUUAUUUCCGGGAUGGCCACAUAAAGGGAGUGUUCCGCUUUUCAGCAGCAGAAA .......((...((..((..((((((...((.((......)).....((((((((((.................))))))))))........)).))))))..))...))...))..... (-25.25 = -26.41 + 1.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:52:37 2011