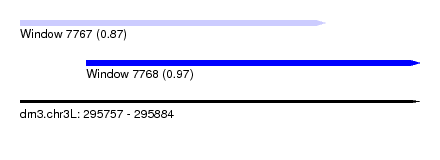
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 295,757 – 295,884 |
| Length | 127 |
| Max. P | 0.973323 |

| Location | 295,757 – 295,854 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 76.03 |
| Shannon entropy | 0.41446 |
| G+C content | 0.38996 |
| Mean single sequence MFE | -27.89 |
| Consensus MFE | -9.89 |
| Energy contribution | -10.37 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.873045 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 295757 97 + 24543557 ---------AGUUUGGUUGGGACUUAAAUUCCGGAAGAAAUGUGU---GUGUAUUUGCGCAAAUUGCAGCGAAAGCU-----GUAAUUAGAGACU----CUCUCUUGAUACCAAUACA ---------.(((((((.((((......))))(..(((..(((((---(......))))))(((((((((....)))-----))))))......)----))..).....))))).)). ( -28.90, z-score = -2.86, R) >droAna3.scaffold_13337 2254473 108 + 23293914 ---------AGACCGUGCGAAAGUUGGGUUCCGCGACAAAUGCGUUCGCUUUUUUUACGUAAAGCGAAGCAUAAUCCAGCUGGUAAUUACCAGCUAGUACUCUCUCUCC-CCAAGACA ---------(((..((((....(((((....).))))..((((.((((((((........)))))))))))).....(((((((....))))))).))))..)))....-........ ( -32.70, z-score = -2.68, R) >droEre2.scaffold_4784 288163 96 + 25762168 ----------CUUUGGUGGGGAAUUUAAUUCCGGAAGAUAUGAGU---CUAUCUUUACGCAAAUUGCAGCGAAAGCU-----GUAAUUACAGACU----CUCUCUUAAUACCAAUACA ----------..((((((.(((((...)))))..((((...((((---((...........(((((((((....)))-----))))))..)))))----)..))))..)))))).... ( -30.32, z-score = -4.57, R) >droYak2.chr3L 274697 106 + 24197627 CUUUUUCUUACUUUGGUUGGGACUUAAAUUCCGGAAGAUAUGUGU---CUAUUUUUACGCAAUUUGCAGCGAAAGCU-----GUAAUUACAGACU----CUCUCUUAAUACCCAUACA ...............((((((..........(....)...(((((---........)))))..(((((((....)))-----)))).........----...........)))).)). ( -22.40, z-score = -1.92, R) >droSec1.super_2 327354 97 + 7591821 ---------AGUUUGCUUGGGACUUAAAUUCCGGAAGAAAUGUGU---CUGUUUUUACGCAAAUUGCAGCGAAAGCU-----GUAAUUAGAGACU----CUCUCUUAAUACCAAUACA ---------....(((...((((....(((.(....).)))..))---))((....)))))(((((((((....)))-----))))))((((...----..))))............. ( -24.90, z-score = -2.20, R) >droSim1.chr2R 8519410 97 + 19596830 ---------AGUUUACUUGGGACUUAAAUUCCGGAAGAGAUGUGU---CUGUUUUUACGCAAAUUGCAGCGAAAGCU-----GUAAUUAGAGACU----CUCUCUUAAUACCAAUACA ---------.......((((((......))))))((((((.(.((---((((....))...(((((((((....)))-----))))))..)))).----)))))))............ ( -28.10, z-score = -3.23, R) >consensus _________AGUUUGGUUGGGACUUAAAUUCCGGAAGAAAUGUGU___CUGUUUUUACGCAAAUUGCAGCGAAAGCU_____GUAAUUACAGACU____CUCUCUUAAUACCAAUACA ................((((((......))))))((((..(((((...........)))))(((((((((....))).....))))))..............))))............ ( -9.89 = -10.37 + 0.48)
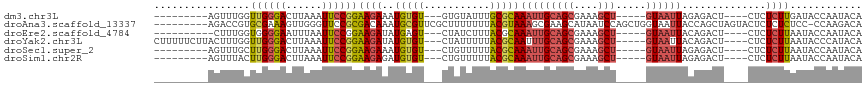
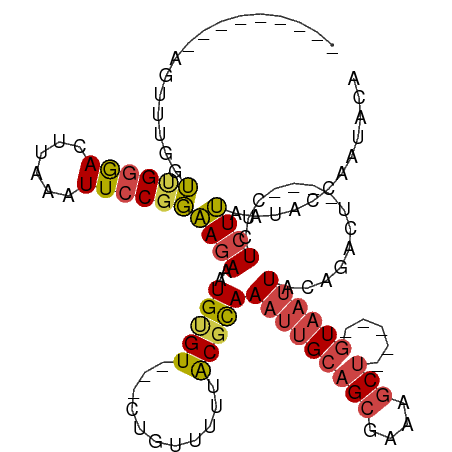
| Location | 295,778 – 295,884 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 75.89 |
| Shannon entropy | 0.44344 |
| G+C content | 0.38025 |
| Mean single sequence MFE | -26.58 |
| Consensus MFE | -12.04 |
| Energy contribution | -13.18 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.973323 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 295778 106 + 24543557 CCGGAAGAAAUGUGU---GUGUAUUUGCGCAAAUUGCAGCGAAAGCUGUAAUUAGAGACUCUCUCUUGAUACCAAUACAAGUCAAAAUGGCAUUUAUAAUGAGGUAAAG----- (((...((..(((((---..(((((......(((((((((....)))))))))((((.....)))).)))))..)))))..))....)))..(((((......))))).----- ( -26.80, z-score = -2.18, R) >droAna3.scaffold_13337 2254494 111 + 23293914 CCGCGACAAAUGCGUUCGCUUUUUUUACGUAAAGCGAAGCAUAAUCC--AGCUGGUAAUUACCAGCUAGUACUCUCUCUCCCCAAGACA-CAUUCGCAUUCAAAUUCAAAUGCA ..((((...((((.((((((((........)))))))))))).....--(((((((....)))))))......................-...))))................. ( -30.00, z-score = -4.70, R) >droEre2.scaffold_4784 288183 106 + 25762168 CCGGAAGAUAUGAGU---CUAUCUUUACGCAAAUUGCAGCGAAAGCUGUAAUUACAGACUCUCUCUUAAUACCAAUACACGUCAAUAUGUUAUUUGUAAUGAGGCAAAG----- ...(((((((.....---.)))))))..((.(((((((((....)))))))))...........((((.(((.((((.((((....)))))))).))).))))))....----- ( -27.50, z-score = -3.17, R) >droYak2.chr3L 274727 106 + 24197627 CCGGAAGAUAUGUGU---CUAUUUUUACGCAAUUUGCAGCGAAAGCUGUAAUUACAGACUCUCUCUUAAUACCCAUACACGUCAAUAGGUUAUUCAUAAUGAGGCAAAG----- ...(((((((.....---.)))))))..((...(((((((....))))))).......(((......((((.((.............)).))))......)))))....----- ( -20.22, z-score = -0.69, R) >droSec1.super_2 327375 106 + 7591821 CCGGAAGAAAUGUGU---CUGUUUUUACGCAAAUUGCAGCGAAAGCUGUAAUUAGAGACUCUCUCUUAAUACCAAUACAAGUCAAAAUGGCAUUUAUAAUGAGGUAAAG----- ((.........((((---(.(((((.((...(((((((((....)))))))))((((.....))))..............)).)))))))))).........)).....----- ( -25.87, z-score = -2.46, R) >droSim1.chr2R 8519431 106 + 19596830 CCGGAAGAGAUGUGU---CUGUUUUUACGCAAAUUGCAGCGAAAGCUGUAAUUAGAGACUCUCUCUUAAUACCAAUACAAGUCAAAAUGGCAUUUAUAAUGAGGUAAAG----- ....((((((.(.((---((((....))...(((((((((....)))))))))..)))).)))))))..((((.......(((.....))).((......))))))...----- ( -29.10, z-score = -3.21, R) >consensus CCGGAAGAAAUGUGU___CUGUUUUUACGCAAAUUGCAGCGAAAGCUGUAAUUAGAGACUCUCUCUUAAUACCAAUACAAGUCAAAAUGGCAUUUAUAAUGAGGUAAAG_____ ..((......(((((...........)))))(((((((((....)))))))))..................))......................................... (-12.04 = -13.18 + 1.14)

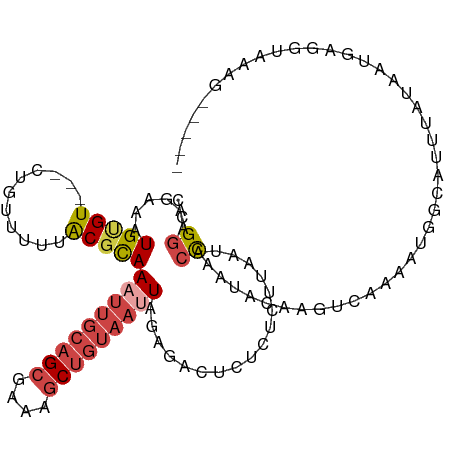
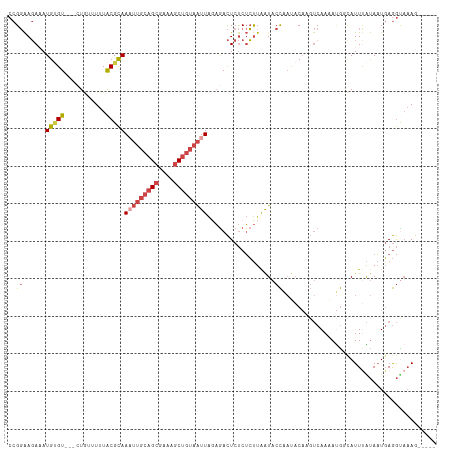
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:52:34 2011