
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 293,118 – 293,226 |
| Length | 108 |
| Max. P | 0.946568 |

| Location | 293,118 – 293,226 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 123 |
| Reading direction | forward |
| Mean pairwise identity | 71.45 |
| Shannon entropy | 0.51760 |
| G+C content | 0.61026 |
| Mean single sequence MFE | -42.45 |
| Consensus MFE | -23.04 |
| Energy contribution | -25.60 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.753925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 293118 108 + 24543557 CGUCGCCUGACGCUGAGCCCCGGGAAGGGGAUGCAGCUGGCGACUCGUCAAUGAGCACCAGUGACGAGUAAGUUGGUUCC-GGUUUCCC-CAAGACGGGCAC--CAUUGGAA----------- .((((((....((((..((((.....))))...)))).))))))((((((.((.....)).))))))......((((.((-.((((...-..)))).)).))--))......----------- ( -45.00, z-score = -1.71, R) >droEre2.scaffold_4784 286079 111 + 25762168 CAACGCCUGACGCUGAGCCCCGGGAAGGGGAUGCAGCUGGCGACUCGUCAAUGAGCACCAGUGACGAGUAAGUUGGUUCC-GGUUCCCCCAAGGACGGGGGCAUCCUUUAAA----------- (((((((....((((..((((.....))))...)))).))).((((((((.((.....)).))))))))..)))).....-((..(((((......)))))...))......----------- ( -49.10, z-score = -2.49, R) >droYak2.chr3L 272368 119 + 24197627 CGACGCCUGACGCGGAGCCCCAGGAAGGGGAUGCGGCUGGCGACUCGUCCACCAGCACCAGUGACGAGUAAGUUGCUUCC-GGUUCCUC-AAGGACGUGGGC--CCCUAGAAUGACCUCCAAA (.((((((...(.((((((((.....))(((.((((((....(((((((.((........))))))))).)))))).)))-)))))).)-.))).))).)..--................... ( -42.30, z-score = 0.29, R) >droSec1.super_2 324915 108 + 7591821 CGUCGCCUGAAGCUGAUCCCCGGGAAGGGGAUGCAGCUGGCGACUCGUCAAUGAGCACCAGUGACGAGUAAGUUGGUUCC-GGUUUCCC-CAAGACGGGCAC--CAUUCGAA----------- .((((((...(((((((((((.....)))))).)))))))))))((((((.((.....)).))))))......((((.((-.((((...-..)))).)).))--))......----------- ( -48.20, z-score = -3.16, R) >droSim1.chr2R 8516777 108 + 19596830 CGUCGCCUGACGCUGAGCCCCGGGAAGGGGAUGCAGCUGGCGACUCGUCAAUGAGCACCAGUGACGAGUAAGUUGGUUCC-GGUGUACC-CAAGACGGGCAC--CAUUCGAA----------- .((((((....((((..((((.....))))...)))).))))))((((((.((.....)).)))))).............-((((..((-(.....))))))--).......----------- ( -44.10, z-score = -1.65, R) >dp4.chrXR_group8 7879685 93 - 9212921 ---CUCCGACAGCUGACAGCCGUCAGGUGGACGAAGCUGCUUCGCCGGCACUGAGCGAAACCGCCAUGUAAGUUGACAGAAGGGUCACCGCACCAC--------------------------- ---....(((.((.....)).))).(((((.(((((...))))))).((((((.(((....))))).))..(.((((......)))).))))))..--------------------------- ( -26.00, z-score = 0.99, R) >consensus CGUCGCCUGACGCUGAGCCCCGGGAAGGGGAUGCAGCUGGCGACUCGUCAAUGAGCACCAGUGACGAGUAAGUUGGUUCC_GGUUCCCC_CAAGACGGGCAC__CAUUCGAA___________ ...((((....((((..((((.....))))...)))).))))((((((((.((.....)).))))))))...................................................... (-23.04 = -25.60 + 2.56)



| Location | 293,118 – 293,226 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 123 |
| Reading direction | reverse |
| Mean pairwise identity | 71.45 |
| Shannon entropy | 0.51760 |
| G+C content | 0.61026 |
| Mean single sequence MFE | -42.99 |
| Consensus MFE | -24.02 |
| Energy contribution | -25.30 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.946568 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 293118 108 - 24543557 -----------UUCCAAUG--GUGCCCGUCUUG-GGGAAACC-GGAACCAACUUACUCGUCACUGGUGCUCAUUGACGAGUCGCCAGCUGCAUCCCCUUCCCGGGGCUCAGCGUCAGGCGACG -----------...(((((--(..((.((..((-((....))-(....)........))..)).))..).)))))....((((((.((((...((((.....))))..))))....)))))). ( -42.10, z-score = -1.81, R) >droEre2.scaffold_4784 286079 111 - 25762168 -----------UUUAAAGGAUGCCCCCGUCCUUGGGGGAACC-GGAACCAACUUACUCGUCACUGGUGCUCAUUGACGAGUCGCCAGCUGCAUCCCCUUCCCGGGGCUCAGCGUCAGGCGUUG -----------......((...((((((....))))))..))-.....((((..((((((((.((.....)).)))))))).(((.((((...((((.....))))..))))....))))))) ( -45.60, z-score = -2.31, R) >droYak2.chr3L 272368 119 - 24197627 UUUGGAGGUCAUUCUAGGG--GCCCACGUCCUU-GAGGAACC-GGAAGCAACUUACUCGUCACUGGUGCUGGUGGACGAGUCGCCAGCCGCAUCCCCUUCCUGGGGCUCCGCGUCAGGCGUCG ....((.(((......(((--((((.((....)-)(((((..-(((.((.(((...........)))((((((((.....)))))))).)).)))..))))).)))))))......))).)). ( -43.60, z-score = 0.68, R) >droSec1.super_2 324915 108 - 7591821 -----------UUCGAAUG--GUGCCCGUCUUG-GGGAAACC-GGAACCAACUUACUCGUCACUGGUGCUCAUUGACGAGUCGCCAGCUGCAUCCCCUUCCCGGGGAUCAGCUUCAGGCGACG -----------......((--((.((.......-((....))-)).))))......((((((.((.....)).))))))(((((((((((.((((((.....)))))))))))...)))))). ( -46.21, z-score = -3.24, R) >droSim1.chr2R 8516777 108 - 19596830 -----------UUCGAAUG--GUGCCCGUCUUG-GGUACACC-GGAACCAACUUACUCGUCACUGGUGCUCAUUGACGAGUCGCCAGCUGCAUCCCCUUCCCGGGGCUCAGCGUCAGGCGACG -----------((((....--((((((.....)-)))))..)-)))..........((((((.((.....)).))))))((((((.((((...((((.....))))..))))....)))))). ( -43.70, z-score = -2.38, R) >dp4.chrXR_group8 7879685 93 + 9212921 ---------------------------GUGGUGCGGUGACCCUUCUGUCAACUUACAUGGCGGUUUCGCUCAGUGCCGGCGAAGCAGCUUCGUCCACCUGACGGCUGUCAGCUGUCGGAG--- ---------------------------((((.((((((((......))))........(((.((((((((.......)))))))).)))))))))))((((((((.....))))))))..--- ( -36.70, z-score = -1.09, R) >consensus ___________UUCGAAUG__GUGCCCGUCUUG_GGGAAACC_GGAACCAACUUACUCGUCACUGGUGCUCAUUGACGAGUCGCCAGCUGCAUCCCCUUCCCGGGGCUCAGCGUCAGGCGACG ............................(((...((....)).)))........((((((((.((.....)).))))))))((((.((((...((((.....))))..))))....))))... (-24.02 = -25.30 + 1.28)

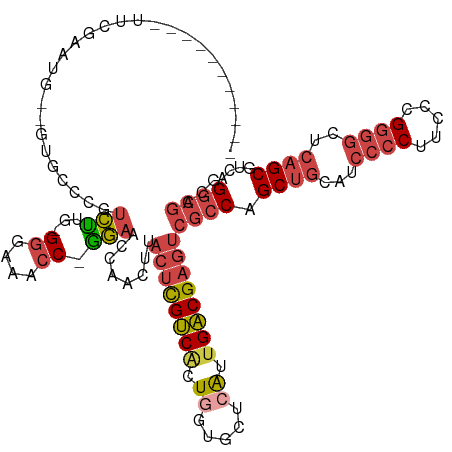
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:52:30 2011