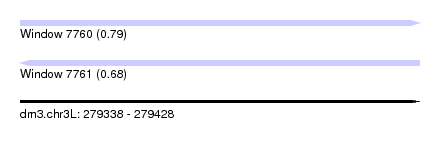
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 279,338 – 279,428 |
| Length | 90 |
| Max. P | 0.788751 |

| Location | 279,338 – 279,428 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 73.78 |
| Shannon entropy | 0.50819 |
| G+C content | 0.57263 |
| Mean single sequence MFE | -29.71 |
| Consensus MFE | -16.57 |
| Energy contribution | -19.45 |
| Covariance contribution | 2.88 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.788751 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
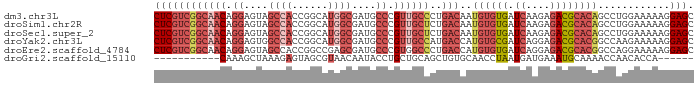
>dm3.chr3L 279338 90 + 24543557 CUCGUCGGCAACAGGAGUAGCCACCGGCAUGGCGAUGCCCGUUGCCCUGACAAUGUGUGAUCAAGAGACGCACAGCCUGGAAAAAGGAGC ...(((((((((.((....((((......))))....)).))))))..)))..((((((.((....)))))))).(((......)))... ( -32.30, z-score = -1.67, R) >droSim1.chr2R 8502935 90 + 19596830 CUCGUCGGCAACAGGAGUAGCCACCGGCAUGGCGAUGCCCGUUGCUCUGACAAUGUGUGAUCAAGAGACGCACAGCCUGGAAAAAGGAGC ...((((....).((((((((....(((((....))))).)))))))))))..((((((.((....)))))))).(((......)))... ( -30.70, z-score = -1.11, R) >droSec1.super_2 310825 90 + 7591821 CUCGUCGGCAACAGGAGUAGCCACCGGCAUGGCGAUGCCCGUUGCUCUGACAAUGUGUGAUCAAGAGACGCACAGCCUGGAAAAAGGAGC ...((((....).((((((((....(((((....))))).)))))))))))..((((((.((....)))))))).(((......)))... ( -30.70, z-score = -1.11, R) >droYak2.chr3L 258064 90 + 24197627 CUCGUCGGCAACAGGAGUGGCCACCGGCAUGGCGAUGCCCGUUGCCAUGACCAUGUGCGAUCAGGAGACGCACGGCCAAGAAAAAGGAGC (((.(.(....)).)))(((((...(((((((((((....))))))))).))..(((((.((....))))))))))))............ ( -38.80, z-score = -2.81, R) >droEre2.scaffold_4784 268531 90 + 25762168 CUCGUCGGCAACAGGAGUAGCCACCGGCCGAGCGAUGCCCGUGGCCCUGACCAUGUGUGAUCAGGAGACGCACGGCCAGGAAAAAGGAGC (((...(((.((....)).))).(((((((.(((..(((...)))(((((.((....)).)))))...))).))))).))......))). ( -35.10, z-score = -1.53, R) >droGri2.scaffold_15110 24449509 73 - 24565398 -----------CAAAGCUAAAGAGUAGCGUAACAAUACCUGCUGCAGCUGUGCAACCUAAUGAUGAAAUGCAAAACCAACACCA------ -----------...((((....(((((.(((....))))))))..)))).((((..............))))............------ ( -10.64, z-score = 0.37, R) >consensus CUCGUCGGCAACAGGAGUAGCCACCGGCAUGGCGAUGCCCGUUGCCCUGACAAUGUGUGAUCAAGAGACGCACAGCCAGGAAAAAGGAGC ((((((((((((.((....((((......))))....)).))))))..)))..((((((.((....))))))))............))). (-16.57 = -19.45 + 2.88)


| Location | 279,338 – 279,428 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 73.78 |
| Shannon entropy | 0.50819 |
| G+C content | 0.57263 |
| Mean single sequence MFE | -30.30 |
| Consensus MFE | -16.42 |
| Energy contribution | -17.40 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.676437 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3L 279338 90 - 24543557 GCUCCUUUUUCCAGGCUGUGCGUCUCUUGAUCACACAUUGUCAGGGCAACGGGCAUCGCCAUGCCGGUGGCUACUCCUGUUGCCGACGAG ...(((......))).((((.(((....)))))))..(((((..(((((((((....(((((....)))))....)))))))))))))). ( -31.40, z-score = -1.21, R) >droSim1.chr2R 8502935 90 - 19596830 GCUCCUUUUUCCAGGCUGUGCGUCUCUUGAUCACACAUUGUCAGAGCAACGGGCAUCGCCAUGCCGGUGGCUACUCCUGUUGCCGACGAG ...(((......))).((((.(((....)))))))..(((((.(.((((((((....(((((....)))))....)))))))))))))). ( -29.00, z-score = -0.85, R) >droSec1.super_2 310825 90 - 7591821 GCUCCUUUUUCCAGGCUGUGCGUCUCUUGAUCACACAUUGUCAGAGCAACGGGCAUCGCCAUGCCGGUGGCUACUCCUGUUGCCGACGAG ...(((......))).((((.(((....)))))))..(((((.(.((((((((....(((((....)))))....)))))))))))))). ( -29.00, z-score = -0.85, R) >droYak2.chr3L 258064 90 - 24197627 GCUCCUUUUUCUUGGCCGUGCGUCUCCUGAUCGCACAUGGUCAUGGCAACGGGCAUCGCCAUGCCGGUGGCCACUCCUGUUGCCGACGAG .(((........((((((((((((....)).)))))..)))))((((((((((....(((((....)))))....))))))))))..))) ( -38.70, z-score = -2.86, R) >droEre2.scaffold_4784 268531 90 - 25762168 GCUCCUUUUUCCUGGCCGUGCGUCUCCUGAUCACACAUGGUCAGGGCCACGGGCAUCGCUCGGCCGGUGGCUACUCCUGUUGCCGACGAG .(((.......(((((((.(((...((((((((....))))))))(((...)))..))).)))))))((((.((....)).))))..))) ( -39.80, z-score = -2.72, R) >droGri2.scaffold_15110 24449509 73 + 24565398 ------UGGUGUUGGUUUUGCAUUUCAUCAUUAGGUUGCACAGCUGCAGCAGGUAUUGUUACGCUACUCUUUAGCUUUG----------- ------(((((.((......))...)))))....((((((....))))))..(((....)))((((.....))))....----------- ( -13.90, z-score = 1.02, R) >consensus GCUCCUUUUUCCAGGCUGUGCGUCUCUUGAUCACACAUUGUCAGGGCAACGGGCAUCGCCAUGCCGGUGGCUACUCCUGUUGCCGACGAG .................(((.(((....)))))).....(((...((((((((....(((((....)))))....)))))))).)))... (-16.42 = -17.40 + 0.98)

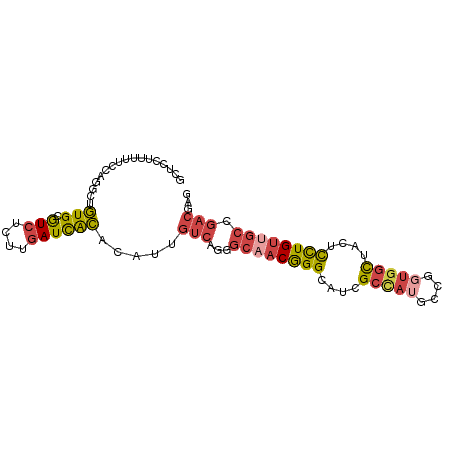
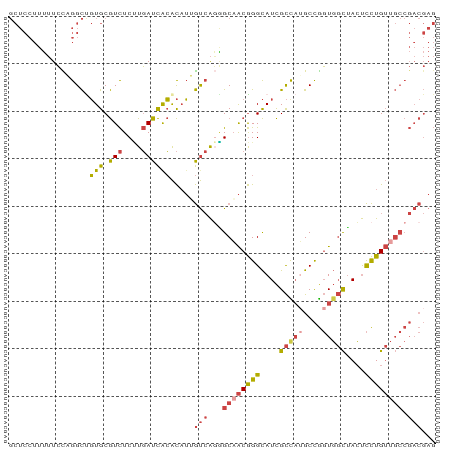
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:52:28 2011