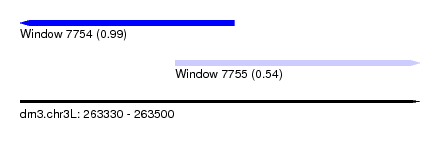
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 263,330 – 263,500 |
| Length | 170 |
| Max. P | 0.988058 |

| Location | 263,330 – 263,421 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 78.99 |
| Shannon entropy | 0.38199 |
| G+C content | 0.38938 |
| Mean single sequence MFE | -19.30 |
| Consensus MFE | -13.46 |
| Energy contribution | -14.58 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.988058 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 263330 91 - 24543557 UUCAUUCAACCUUGCAUUUGGAUGAUUCCCAUUUUC--AAAAAAACCUGUUGCUUUUACAGGUAAUAUCUGACGCCCUUAUCAGUAACACCUU--- .((((((((........))))))))...........--......((((((.......)))))).....((((........)))).........--- ( -15.40, z-score = -1.86, R) >droSim1.chr2R 8487460 94 - 19596830 UUCAUUCAGCCUUGCGUUUGGAUGACUCCCAUUUUC--AAAAAAACCUGUUGCUUUUACAGGUAAUAGCUGACGCCCUUAUCAGCAACACUUAUUU .((((((((.(....).))))))))...........--......((((((.......))))))....(((((........)))))........... ( -19.70, z-score = -2.01, R) >droSec1.super_2 290780 96 - 7591821 UUCAUUCAGCCUUGCAUUUGGAUGACUCCCAUUUUCAAAAAAAAACCUGUUGCUUUUACAGGUAAUAGCUGACGCCCUUAUCAGCAACACUUAUUU .((((((((........))))))))...................((((((.......))))))....(((((........)))))........... ( -19.60, z-score = -2.20, R) >droYak2.chr3L 241815 77 - 24197627 UUCAUUCAGCCUUGC---UGAAUG---------------AAAAAACCUGUUGAUUUAACAGGUAAUAGCUGACGCCCUC-UAAGCGACACUUACUU ((((((((((...))---))))))---------------))...((((((((...))))))))..(((.((.(((....-...))).)).)))... ( -22.80, z-score = -4.78, R) >droEre2.scaffold_4784 252009 92 - 25762168 UUCAUUCAGUC-UGCUCUUGGACGUUGUCCCAAUU---GAAAAAACCUGUUGAUUGAACAGGUAAUAGCUGACGCCCUCAUAAGCAACACUAACUU ........(((-.(((...((((...)))).....---......(((((((.....)))))))...))).)))((........))........... ( -19.00, z-score = -1.14, R) >consensus UUCAUUCAGCCUUGCAUUUGGAUGACUCCCAUUUUC__AAAAAAACCUGUUGCUUUUACAGGUAAUAGCUGACGCCCUUAUCAGCAACACUUACUU .((((((((........))))))))...................((((((.......))))))....(((((........)))))........... (-13.46 = -14.58 + 1.12)
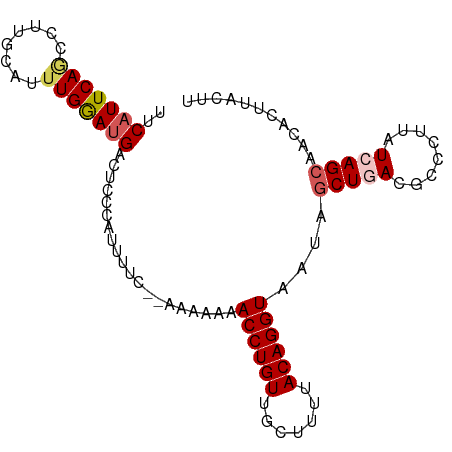
| Location | 263,396 – 263,500 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 83.51 |
| Shannon entropy | 0.30320 |
| G+C content | 0.47410 |
| Mean single sequence MFE | -27.53 |
| Consensus MFE | -22.58 |
| Energy contribution | -22.94 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.538264 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 263396 104 + 24543557 UCAUCCAAAUGCAAGGUUGAAUGAAUUUCGGGAUGUUUUCUUUGGUUGCAGGGUGCCAACGUCACCAAA-UCAGAUUACGCCUGUAAAUCUCUACAGCCGCUGGA ...((((...((..(((((...((((((((((.(((..((((((((.((..(....)...)).))))).-..)))..))))))).)))).))..))))))))))) ( -26.40, z-score = -0.22, R) >droSim1.chr2R 8487529 104 + 19596830 UCAUCCAAACGCAAGGCUGAAUGAAUUUCGGGAUGUAUUCUUUGGUUGCAGGGUGCCAACGUCACCAAA-UCAGAUUACGCCCGUAAAUCUCAACAGCCACUGUG .........((((.(((((..(((((((((((.((((...(((((((....(((((....).)))).))-))))).)))))))).)))).))).)))))..)))) ( -32.00, z-score = -2.58, R) >droSec1.super_2 290851 98 + 7591821 UCAUCCAAAUGCAAGGCUGAAUGAAUUUCGGGAUGUAUUCUUUGGUUGCAGGGUGCCAACGUCACCAAA-UCAGAUUACGCCCGUAAAUCUCAACAGCC------ ..............(((((..(((((((((((.((((...(((((((....(((((....).)))).))-))))).)))))))).)))).))).)))))------ ( -28.70, z-score = -2.06, R) >droYak2.chr3L 241880 91 + 24197627 -------------AGGCUGAAUGAAUUUCGGGAUGUAUUCUUUGGUUGCAGGGUGCCAACGUCACCAAA-CCAGAUUGCGCCCGUAAAUCUCUGCAGCCGCUGGG -------------.(((((...((((((((((.((((...(((((((....(((((....).)))).))-))))).)))))))).)))).))..)))))...... ( -33.60, z-score = -2.05, R) >droEre2.scaffold_4784 252077 102 + 25762168 ACGUCCAAGAGC-AGACUGAAUGAAUUUCG--AUGUAUUCUUUGGUUGCAAUGUGCCGACGUCACCAAAACCAGAUUACGCCCGUAAAUCUCUUUAGCCAGUGGG (((((((...((-((.(..((.((((....--....))))))..)))))....))..)))))..(((.....(((((((....)).)))))..........))). ( -16.96, z-score = 1.56, R) >consensus UCAUCCAAAUGCAAGGCUGAAUGAAUUUCGGGAUGUAUUCUUUGGUUGCAGGGUGCCAACGUCACCAAA_UCAGAUUACGCCCGUAAAUCUCUACAGCCACUGGG ..............(((((...((((((((((.((((.((((((((.((..(....)...)).)))))....))).)))))))).)))).))..)))))...... (-22.58 = -22.94 + 0.36)
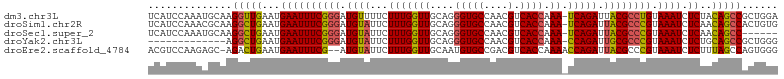

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:52:23 2011