| Sequence ID | dm3.chr3L |
|---|---|
| Location | 261,854 – 261,959 |
| Length | 105 |
| Max. P | 0.999508 |

| Location | 261,854 – 261,959 |
|---|---|
| Length | 105 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 65.02 |
| Shannon entropy | 0.73089 |
| G+C content | 0.47959 |
| Mean single sequence MFE | -36.50 |
| Consensus MFE | -14.05 |
| Energy contribution | -12.56 |
| Covariance contribution | -1.49 |
| Combinations/Pair | 1.60 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.84 |
| SVM RNA-class probability | 0.995735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
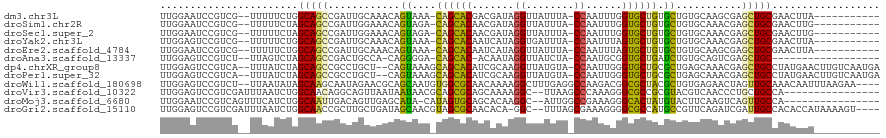
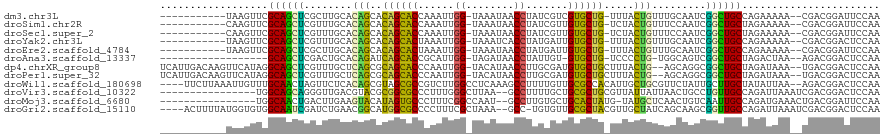
>dm3.chr3L 261854 105 + 24543557 UUGGAAUCCGUCG--UUUUUCUGGCAGCCGAUUGCAAACAGUAAA-CAGCACGACGAUAGGUUAUUUA-CCAAUUUGGUGCUGUGCUGUGCAAGCGAGCUGCGAACUUA----------- ..((((.......--...)))).(((((((.((((..(((((..(-((((((.(.(((.(((.....)-)).)))).)))))))))))))))).)).))))).......----------- ( -36.00, z-score = -2.36, R) >droSim1.chr2R 8485971 105 + 19596830 UUGGAAUCCGUCG--UUUUUCUAGCAGCCGAUUGGAAACAGUAGA-CAGCACAACGAUAGGUUAUUUA-CCAAUUUGGUGCUGUGCUGUGCAAACGAGCUGCGAACUUG----------- .(((((.......--...)))))(((((((.(((...(((((..(-((((((...(((.(((.....)-)).)))..)))))))))))).))).)).))))).......----------- ( -35.20, z-score = -2.79, R) >droSec1.super_2 289293 105 + 7591821 UUGGAAUCCGUCG--UUUUUCUAGCAGCCGAUUGGAAACAGUAGA-CAGCACAACGAUAGGUUAUUUA-CCAAUUUGGUGCUGUGCUGUGCAAACGAGCUGCGAACUUG----------- .(((((.......--...)))))(((((((.(((...(((((..(-((((((...(((.(((.....)-)).)))..)))))))))))).))).)).))))).......----------- ( -35.20, z-score = -2.79, R) >droYak2.chr3L 240316 105 + 24197627 UUGGAGUCCGUCG--UUUUUCUGGCAGCCGAUUGCAAACAGUAAA-CAGCACAAUCAUAGGUGAUUUA-CCAAUUUAGUGCUGUGCUGUGCAAACGAGCUGCGAACUUA----------- ...((((.(..((--......))(((((((.((((..(((((..(-((((((.......((((...))-))......)))))))))))))))).)).)))))).)))).----------- ( -33.02, z-score = -1.83, R) >droEre2.scaffold_4784 250483 105 + 25762168 UUGGAAUCCGUCG--UUUUUCUGGCAGCCGAUUGCAAACAGUAAA-CAGCACAAUCAUAGGUUAUUUA-CCAAUUUAGUGCUGUGCUGUGCAAGCGAGCUGCGAACUUA----------- ..((((.......--...)))).(((((((.((((..(((((..(-((((((.......(((.....)-))......)))))))))))))))).)).))))).......----------- ( -33.62, z-score = -2.28, R) >droAna3.scaffold_13337 2202096 96 + 23293914 UUGGAGUCCGUCU--UUAGUCUAGCAGCCGACUGCCA-CAGGGGA-CAGCAC-ACAAUAGGUUAUCUA-CCAAUGCGGUGCUGAUCUGUGCAGUCGAGCUGC------------------ ((((((.....))--))))....((((((((((((.(-((((...-((((((-...((.(((.....)-)).))...)))))).)))))))))))).)))))------------------ ( -42.80, z-score = -3.85, R) >dp4.chrXR_group8 7840137 115 - 9212921 UUGGAGUCCGUCA--UUUAUCUAGCAGCCGCCUGCU--CAGUAAAGCAGCACAUCGCAAGGUUAUGUA-CCAAUUGGGUGCUGCGCUGAGCAAACGAGCUGCCUAUGAACUUGUCAAUGA ((((......(((--(.......(((((((..((((--((((...((((((((((....)))......-((....)))))))))))))))))..)).)))))..)))).....))))... ( -43.30, z-score = -2.74, R) >droPer1.super_32 103342 115 + 992631 UUGGAGUCCGUCA--UUUAUCUAGCAGCCGCCUGCU--CAGUAAAGCAGCACAUCGCAAGGUUAUGUA-CCAAUUGGGUGCUGCGCUGAGCAAACGAGCUGCCUAUGAACUUGUCAAUGA ((((......(((--(.......(((((((..((((--((((...((((((((((....)))......-((....)))))))))))))))))..)).)))))..)))).....))))... ( -43.30, z-score = -2.74, R) >droWil1.scaffold_180698 50918 114 - 11422946 UUGGAGUCCGUCU--UUAAUAUAGCAAGCAAUAGAACGCAGCAAUGUGGCGCAACAAAAGGCUUUGAGGCCAAGACGGCGCUACGCUGUGAGAACUAGUUGCAAACAAUUUAAGAA---- ((((((.....))--))))........(((((((..((((((...(((((((.......((((....))))......)))))))))))))....)).)))))..............---- ( -32.22, z-score = -1.18, R) >droVir3.scaffold_10322 380019 102 + 456481 UUGGAGUCCGUCGAUUUAAUCUGGCAACAGGCAGUUAAUAAUAACGCAGCGCAGCAAAAGGC--UUAAGCCCAAAGGGCGCCGCGUACGUCAACCCUGCUGCCA---------------- ...(((((....))))).....((((.((((..(((.((....((((.((((.......(((--....)))......)))).))))..)).))))))).)))).---------------- ( -31.52, z-score = -0.54, R) >droMoj3.scaffold_6680 2417329 101 + 24764193 UUGGAAUCCGUCAGUUUCAUCUGGCAAUUGACAGUUGAGCAUA-CAUAGUGCAGCACAAGGC--AUUGGCCGAAAGGGCACUAUGUACUUCAAGUCAGUUGCCA---------------- .((((((......))))))...((((((((((..(((((..((-((((((((.((.....))--.....((....)))))))))))).))))))))))))))).---------------- ( -41.80, z-score = -5.15, R) >droGri2.scaffold_15110 24419480 113 - 24565398 UUGGAGUCCGUCGAUUUAAUCUGGCAACCGCUUGCUGAUAGCAACGUAGCGCAACACA-GGC--UUUAGCGAAAGGGGCGCCAUGCCGUUCAGAUCGAUUGCCACACCAUAAAAGU---- .(((.(((.((((((((....(((((..((.((((.....))))))..((((..(...-.((--....))....)..))))..)))))...)))))))).)..)).))).......---- ( -30.00, z-score = 0.49, R) >consensus UUGGAGUCCGUCG__UUUAUCUAGCAGCCGAUUGCAAACAGUAAA_CAGCACAACAAAAGGUUAUUUA_CCAAUUGGGUGCUGUGCUGUGCAAACGAGCUGCCAACUUA___________ .......................(((((............((....((((((.......((........))......)))))).))...........))))).................. (-14.05 = -12.56 + -1.49)
| Location | 261,854 – 261,959 |
|---|---|
| Length | 105 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 65.02 |
| Shannon entropy | 0.73089 |
| G+C content | 0.47959 |
| Mean single sequence MFE | -35.21 |
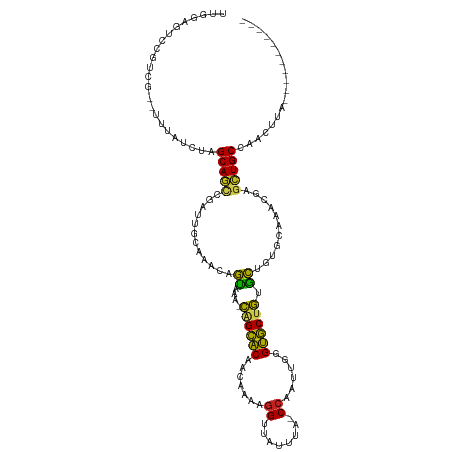
| Consensus MFE | -16.95 |
| Energy contribution | -15.07 |
| Covariance contribution | -1.88 |
| Combinations/Pair | 1.82 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.96 |
| SVM RNA-class probability | 0.999508 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 261854 105 - 24543557 -----------UAAGUUCGCAGCUCGCUUGCACAGCACAGCACCAAAUUGG-UAAAUAACCUAUCGUCGUGCUG-UUUACUGUUUGCAAUCGGCUGCCAGAAAAA--CGACGGAUUCCAA -----------.......(((((.((.((((((((.(((((((......((-(.....))).......))))))-)...))))..)))).)))))))........--............. ( -30.92, z-score = -1.96, R) >droSim1.chr2R 8485971 105 - 19596830 -----------CAAGUUCGCAGCUCGUUUGCACAGCACAGCACCAAAUUGG-UAAAUAACCUAUCGUUGUGCUG-UCUACUGUUUCCAAUCGGCUGCUAGAAAAA--CGACGGAUUCCAA -----------.......(((((.((.(((.((((.(((((((......((-(.....))).......))))))-)...))))...))).)))))))........--............. ( -27.92, z-score = -1.61, R) >droSec1.super_2 289293 105 - 7591821 -----------CAAGUUCGCAGCUCGUUUGCACAGCACAGCACCAAAUUGG-UAAAUAACCUAUCGUUGUGCUG-UCUACUGUUUCCAAUCGGCUGCUAGAAAAA--CGACGGAUUCCAA -----------.......(((((.((.(((.((((.(((((((......((-(.....))).......))))))-)...))))...))).)))))))........--............. ( -27.92, z-score = -1.61, R) >droYak2.chr3L 240316 105 - 24197627 -----------UAAGUUCGCAGCUCGUUUGCACAGCACAGCACUAAAUUGG-UAAAUCACCUAUGAUUGUGCUG-UUUACUGUUUGCAAUCGGCUGCCAGAAAAA--CGACGGACUCCAA -----------..((((((((((.((.((((((((.(((((((...((.((-(.....))).))....))))))-)...))))..)))).)))))))..(.....--)...))))).... ( -33.60, z-score = -2.77, R) >droEre2.scaffold_4784 250483 105 - 25762168 -----------UAAGUUCGCAGCUCGCUUGCACAGCACAGCACUAAAUUGG-UAAAUAACCUAUGAUUGUGCUG-UUUACUGUUUGCAAUCGGCUGCCAGAAAAA--CGACGGAUUCCAA -----------.......(((((.((.((((((((.(((((((...((.((-(.....))).))....))))))-)...))))..)))).)))))))........--............. ( -31.90, z-score = -2.33, R) >droAna3.scaffold_13337 2202096 96 - 23293914 ------------------GCAGCUCGACUGCACAGAUCAGCACCGCAUUGG-UAGAUAACCUAUUGU-GUGCUG-UCCCCUG-UGGCAGUCGGCUGCUAGACUAA--AGACGGACUCCAA ------------------(((((.(((((((((((..((((((.(((..((-(.....)))...)))-))))))-....)))-).))))))))))))........--............. ( -41.60, z-score = -4.12, R) >dp4.chrXR_group8 7840137 115 + 9212921 UCAUUGACAAGUUCAUAGGCAGCUCGUUUGCUCAGCGCAGCACCCAAUUGG-UACAUAACCUUGCGAUGUGCUGCUUUACUG--AGCAGGCGGCUGCUAGAUAAA--UGACGGACUCCAA .........(((((...((((((.(((((((((((.(((((((......((-(.....))).......)))))))....)))--)))))))))))))).(.....--...)))))).... ( -44.92, z-score = -3.50, R) >droPer1.super_32 103342 115 - 992631 UCAUUGACAAGUUCAUAGGCAGCUCGUUUGCUCAGCGCAGCACCCAAUUGG-UACAUAACCUUGCGAUGUGCUGCUUUACUG--AGCAGGCGGCUGCUAGAUAAA--UGACGGACUCCAA .........(((((...((((((.(((((((((((.(((((((......((-(.....))).......)))))))....)))--)))))))))))))).(.....--...)))))).... ( -44.92, z-score = -3.50, R) >droWil1.scaffold_180698 50918 114 + 11422946 ----UUCUUAAAUUGUUUGCAACUAGUUCUCACAGCGUAGCGCCGUCUUGGCCUCAAAGCCUUUUGUUGCGCCACAUUGCUGCGUUCUAUUGCUUGCUAUAUUAA--AGACGGACUCCAA ----.((((.(((.((..((((.(((....(.((((((.((((......(((......))).......)))).))...)))).)..)))))))..))...))).)--))).......... ( -26.42, z-score = -0.37, R) >droVir3.scaffold_10322 380019 102 - 456481 ----------------UGGCAGCAGGGUUGACGUACGCGGCGCCCUUUGGGCUUAA--GCCUUUUGCUGCGCUGCGUUAUUAUUAACUGCCUGUUGCCAGAUUAAAUCGACGGACUCCAA ----------------(((((((((((((((...(((((((((.....((((....--))))......))))))))).....)))))..))))))))))..................... ( -42.90, z-score = -3.35, R) >droMoj3.scaffold_6680 2417329 101 - 24764193 ----------------UGGCAACUGACUUGAAGUACAUAGUGCCCUUUCGGCCAAU--GCCUUGUGCUGCACUAUG-UAUGCUCAACUGUCAAUUGCCAGAUGAAACUGACGGAUUCCAA ----------------((((((.((((((((.(((((((((((......(((....--))).......))))))))-)))..))))..)))).))))))..................... ( -34.92, z-score = -4.08, R) >droGri2.scaffold_15110 24419480 113 + 24565398 ----ACUUUUAUGGUGUGGCAAUCGAUCUGAACGGCAUGGCGCCCCUUUCGCUAAA--GCC-UGUGUUGCGCUACGUUGCUAUCAGCAAGCGGUUGCCAGAUUAAAUCGACGGACUCCAA ----.......((((.(((((((((..((((.((((.((((((......(((....--...-.)))..)))))).))))...))))....))))))))).))))................ ( -34.60, z-score = -0.45, R) >consensus ___________UAAGUUCGCAGCUCGUUUGCACAGCACAGCACCAAAUUGG_UAAAUAACCUAUCGUUGUGCUG_UUUACUGUUUGCAAUCGGCUGCCAGAUAAA__CGACGGACUCCAA ..................((((((........(((..((((((......((........)).......)))))).....))).........))))))....................... (-16.95 = -15.07 + -1.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:52:22 2011