| Sequence ID | dm3.chr3L |
|---|---|
| Location | 260,191 – 260,298 |
| Length | 107 |
| Max. P | 0.687862 |

| Location | 260,191 – 260,291 |
|---|---|
| Length | 100 |
| Sequences | 13 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 71.87 |
| Shannon entropy | 0.59478 |
| G+C content | 0.41743 |
| Mean single sequence MFE | -19.98 |
| Consensus MFE | -14.04 |
| Energy contribution | -13.42 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.28 |
| Mean z-score | -0.12 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.525776 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
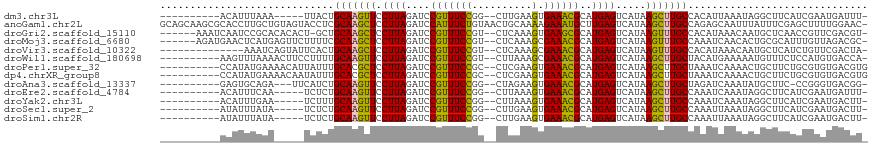
>dm3.chr3L 260191 100 - 24543557 ----------ACAUUUAAA-----UUACUGCAAGUUCCUUAGAUCCGUUUCCGG--CUUGAAGUGAAACGCAUGAGUCAUAAGCUUGCCACAUUAAAUAGGCUUCAUCGAAUGAUUU- ----------.........-----....(((...((((((.((.(((....)))--.)).))).)))..))).((((((((((((((..........)))))))......)))))))- ( -16.10, z-score = 0.60, R) >anoGam1.chr2L 17967602 117 - 48795086 GCAGCAAGCGCACCUUGCUGUAGUACCUCGCAAGCUCCUUAGAUCCAUUUCUGUAACUGCAAAAGAAAUGCUUGAGUCAUAAGCUUGCCAGAGCAAUUUAUUUCGAGCUUUUGGAAC- ((((((((.....))))))))....((....(((((((((((...(((((((...........))))))).)))))......((((....))))..........))))))..))...- ( -30.50, z-score = -0.67, R) >droGri2.scaffold_15110 24417780 108 + 24565398 ------AAAUCAAUCCGCACACACU-GCUGCAAGCUCCUUAGAUCCGUUUCCGU--CUCAAAGUGAAGCGCAUGAGUCAUAAGUUUGCCACAUAAACAAUGCUCAACCGUUCGACGU- ------.........((.((.....-..(((..(((((((.((..((....)).--.)).))).).))))))((((.(((..(((((.....))))).)))))))...)).))....- ( -16.60, z-score = 0.87, R) >droMoj3.scaffold_6680 2415653 109 - 24764193 ------AGAUGAACUCAUGAGUUCUUUUCGCAAGCUCCUUAGAUCCGUUUCCGU--CUCAAAGCGAAACGCAUGAGUCAUAAGUUUGCCAAAUCAACACUGCGCAUUUGUUAGACGC- ------..((((.((((((.(((....(((((((...))).((..((....)).--.))...))))))).))))))))))..(((((.((((((........).))))).)))))..- ( -23.20, z-score = -0.19, R) >droVir3.scaffold_10322 378344 101 - 456481 --------------AAAUCAGUAUUCACUGCAAGCUCCUUAGAUCCGUUUCCGU--CUCAAAGCGAAACGCAUGAGUCAUAAGUUUGCCACAUAAACAAUGCUCAUCUGUUCGACUA- --------------....((((....))))...........((..((....)).--.))..((((((.((.(((((.(((..(((((.....))))).)))))))).)))))).)).- ( -15.90, z-score = -0.15, R) >droWil1.scaffold_180698 49167 105 + 11422946 ----------AAGUUUAAAACUUCCUUUUGCAAGUUCCUUAGAUCCGUUUCCGU--CUUAAAGCGAAACGCAUGAGUCAUAAGCUUGCUACAUGAAAAAUGUUUCUCCAUGUGACCA- ----------...................(((((((.((((....(((((((..--......).))))))..)))).....)))))))((((((.............))))))....- ( -18.42, z-score = -1.08, R) >droPer1.super_32 101654 106 - 992631 ----------CCAUAUGAAAACAUUAUUUGCACGCUCCUUAGAUCCGUUUCCGC--CUCGAAGUGAAACGCAUGAGUCAUAAGCUUGCUAAAUCAAAACUGCUUCUGCGUGUGACGUG ----------.................(..(((((..((((....(((((((..--......).))))))..))))....((((................))))..)))))..).... ( -19.59, z-score = 0.04, R) >dp4.chrXR_group8 7838449 106 + 9212921 ----------CCAUAUGAAAACAAUAUUUGCACGCUCCUUAGAUCCGUUUCCGC--CUCGAAGUGAAACGCAUGAGUCAUAAGCUUGCUAAAUCAAAACUGCUUCUGCGUGUGACGUG ----------.................(..(((((..((((....(((((((..--......).))))))..))))....((((................))))..)))))..).... ( -19.59, z-score = -0.06, R) >droAna3.scaffold_13337 2200413 101 - 23293914 ----------GAGUGCAGA---UUCAUCUGCAAGUUCCUUAGAUCCGUUUCCGG--CUAGAAGUGAAACGCAUGAGUCAUAAGCUUGCUAGAUCAAAUAUGCUUC-CCGGGUGACGG- ----------(((.(((.(---((.(((((((((((.((((....(((((((..--......).))))))..)))).....))))))).))))..))).))))))-(((.....)))- ( -24.50, z-score = 0.11, R) >droEre2.scaffold_4784 248813 100 - 25762168 ----------ACAUUUCAA-----UCUCUGCAAGUUCCUUAGAUCCGUUUCCGG--CUUAAAGUGAAACGCAUGAGUCAUAAGCUUGCCAAAUCAAAUAGGCUUCAUCGAAUGAUUU- ----------.(((((...-----.((((((...((((((.((.(((....)))--.)).))).)))..))).)))....(((((((..........)))))))....)))))....- ( -18.30, z-score = -0.31, R) >droYak2.chr3L 238653 100 - 24197627 ----------ACAUUUGAA-----UCUUUGCAAGUUCCUUAGAUCCGUUUCCGG--CUUAAAGUGAAACGCAUGAGUCAUAAGCUUGCCAAAUCAAAUAGGCUUCAUCGAAUGACUU- ----------.(((((((.-----((..(((...((((((.((.(((....)))--.)).))).)))..))).)).....(((((((..........)))))))..)))))))....- ( -20.30, z-score = -0.62, R) >droSec1.super_2 287623 100 - 7591821 ----------AUAUUUAUA-----UCUCUGCAAGUUCCUUAGAUCCGUUUCCGG--CUUGAAGUGAAACGCAUGAGUCAUAAGCUUGCCAAAUUAAAUAGGCUUCAUCGAAUGACUU- ----------.........-----....(((...((((((.((.(((....)))--.)).))).)))..))).((((((((((((((..........)))))))......)))))))- ( -18.40, z-score = -0.08, R) >droSim1.chr2R 8484301 100 - 19596830 ----------AUAUUUAUA-----UCUCUGCAAGUUCCUUAGAUCCGUUUCCGG--CUUGAAGUGAAACGCAUGAGUCAUAAGCUUGCCAAAUUAAAUAGGCUUCAUCGAAUGACUU- ----------.........-----....(((...((((((.((.(((....)))--.)).))).)))..))).((((((((((((((..........)))))))......)))))))- ( -18.40, z-score = -0.08, R) >consensus __________AAAUUUAAA_____UCUCUGCAAGUUCCUUAGAUCCGUUUCCGG__CUUAAAGUGAAACGCAUGAGUCAUAAGCUUGCCAAAUCAAAUAUGCUUCAUCGUAUGACUU_ .............................(((((((.((((....(((((((..........).))))))..)))).....))))))).............................. (-14.04 = -13.42 + -0.63)


| Location | 260,206 – 260,298 |
|---|---|
| Length | 92 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.13 |
| Shannon entropy | 0.56011 |
| G+C content | 0.41170 |
| Mean single sequence MFE | -17.90 |
| Consensus MFE | -14.04 |
| Energy contribution | -13.42 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.28 |
| Mean z-score | -0.29 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.687862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
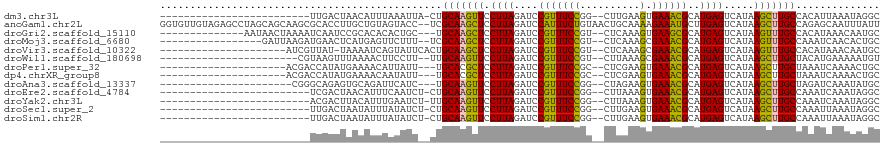
>dm3.chr3L 260206 92 - 24543557 -------------------------UUGACUAACAUUUAAAUUA-CUGCAAGUUCCUUAGAUCCGUUUCCGG--CUUGAAGUGAAACGCAUGAGUCAUAAGCUUGCCACAUUAAAUAGGC -------------------------.........((((((....-..(((((((.((((....(((((((..--......).))))))..)))).....)))))))....)))))).... ( -15.00, z-score = 0.43, R) >anoGam1.chr2L 17967617 118 - 48795086 GGUGUUGUAGAGCCUAGCAGCAAGCGCACCUUGCUGUAGUACC--UCGCAAGCUCCUUAGAUCCAUUUCUGUAACUGCAAAAGAAAUGCUUGAGUCAUAAGCUUGCCAGAGCAAUUUAUU ...(((((.(((.(((.(((((((.....))))))))))...)--))(((((((.(((((...(((((((...........))))))).))))).....)))))))....)))))..... ( -32.20, z-score = -0.66, R) >droGri2.scaffold_15110 24417795 101 + 24565398 --------------AAUAACUAAAAUCAAUCCGCACACACUGC---UGCAAGCUCCUUAGAUCCGUUUCCGU--CUCAAAGUGAAGCGCAUGAGUCAUAAGUUUGCCACAUAAACAAUGC --------------..................(((.....)))---.(((((((.((((....(((((((..--......).))))))..)))).....))))))).............. ( -16.00, z-score = -0.15, R) >droMoj3.scaffold_6680 2415668 99 - 24764193 -----------------GAUUAAGAUGAACUCAUGAGUUCUUU--UCGCAAGCUCCUUAGAUCCGUUUCCGU--CUCAAAGCGAAACGCAUGAGUCAUAAGUUUGCCAAAUCAACACUGC -----------------((((..((.(((((....)))))...--))(((((((.((((....(((((((..--......).))))))..)))).....)))))))..))))........ ( -22.20, z-score = -1.50, R) >droVir3.scaffold_10322 378359 96 - 456481 ---------------------AUCGUUAU-UAAAAUCAGUAUUCACUGCAAGCUCCUUAGAUCCGUUUCCGU--CUCAAAGCGAAACGCAUGAGUCAUAAGUUUGCCACAUAAACAAUGC ---------------------...(((..-.......(((....)))(((((((.((((....(((((((..--......).))))))..)))).....)))))))......)))..... ( -14.90, z-score = -0.69, R) >droWil1.scaffold_180698 49182 93 + 11422946 -----------------------CGUAAGUUUAAAACUUCCUU--UUGCAAGUUCCUUAGAUCCGUUUCCGU--CUUAAAGCGAAACGCAUGAGUCAUAAGCUUGCUACAUGAAAAAUGU -----------------------.(((((((((.(((((.(..--..).))))).((((....(((((((..--......).))))))..))))...))))))))).((((.....)))) ( -19.10, z-score = -1.92, R) >droPer1.super_32 101670 94 - 992631 ---------------------ACGACCAUAUGAAAACAUUAUU---UGCACGCUCCUUAGAUCCGUUUCCGC--CUCGAAGUGAAACGCAUGAGUCAUAAGCUUGCUAAAUCAAAACUGC ---------------------......................---.(((.(((.((((....(((((((..--......).))))))..)))).....))).))).............. ( -13.10, z-score = 0.52, R) >dp4.chrXR_group8 7838465 94 + 9212921 ---------------------ACGACCAUAUGAAAACAAUAUU---UGCACGCUCCUUAGAUCCGUUUCCGC--CUCGAAGUGAAACGCAUGAGUCAUAAGCUUGCUAAAUCAAAACUGC ---------------------......................---.(((.(((.((((....(((((((..--......).))))))..)))).....))).))).............. ( -13.10, z-score = 0.42, R) >droAna3.scaffold_13337 2200427 93 - 23293914 ----------------------CGGGCAGAGUGCAGAUUCAUC---UGCAAGUUCCUUAGAUCCGUUUCCGG--CUAGAAGUGAAACGCAUGAGUCAUAAGCUUGCUAGAUCAAAUAUGC ----------------------.(((.((..((((((....))---))))..)))))..((((.((((((..--......).)))))(((..(((.....))))))..))))........ ( -22.10, z-score = 0.08, R) >droEre2.scaffold_4784 248828 92 - 25762168 -------------------------UCGACUAACAUUUCAAUCU-CUGCAAGUUCCUUAGAUCCGUUUCCGG--CUUAAAGUGAAACGCAUGAGUCAUAAGCUUGCCAAAUCAAAUAGGC -------------------------...................-..(((((((.((((....(((((((..--......).))))))..)))).....))))))).............. ( -14.40, z-score = 0.30, R) >droYak2.chr3L 238668 92 - 24197627 -------------------------ACGACUUACAUUUGAAUCU-UUGCAAGUUCCUUAGAUCCGUUUCCGG--CUUAAAGUGAAACGCAUGAGUCAUAAGCUUGCCAAAUCAAAUAGGC -------------------------.........((((((...(-(((((((((.((((....(((((((..--......).))))))..)))).....)))))).)))))))))).... ( -17.40, z-score = -0.32, R) >droSec1.super_2 287638 92 - 7591821 -------------------------UUGACUAAUAUUUAUAUCU-CUGCAAGUUCCUUAGAUCCGUUUCCGG--CUUGAAGUGAAACGCAUGAGUCAUAAGCUUGCCAAAUUAAAUAGGC -------------------------..........(((((..((-((((...((((((.((.(((....)))--.)).))).)))..))).)))..)))))...(((..........))) ( -16.60, z-score = -0.15, R) >droSim1.chr2R 8484316 92 - 19596830 -------------------------UUGACUAAUAUUUAUAUCU-CUGCAAGUUCCUUAGAUCCGUUUCCGG--CUUGAAGUGAAACGCAUGAGUCAUAAGCUUGCCAAAUUAAAUAGGC -------------------------..........(((((..((-((((...((((((.((.(((....)))--.)).))).)))..))).)))..)))))...(((..........))) ( -16.60, z-score = -0.15, R) >consensus _________________________UCAACUAACAUUUAUAUC__CUGCAAGUUCCUUAGAUCCGUUUCCGG__CUUAAAGUGAAACGCAUGAGUCAUAAGCUUGCCAAAUCAAAUAUGC ...............................................(((((((.((((....(((((((..........).))))))..)))).....))))))).............. (-14.04 = -13.42 + -0.63)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:52:20 2011