| Sequence ID | dm3.chr3L |
|---|---|
| Location | 251,981 – 252,072 |
| Length | 91 |
| Max. P | 0.904572 |

| Location | 251,981 – 252,072 |
|---|---|
| Length | 91 |
| Sequences | 13 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 70.82 |
| Shannon entropy | 0.64933 |
| G+C content | 0.42725 |
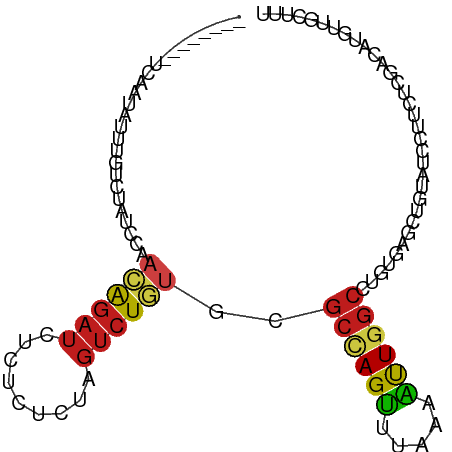
| Mean single sequence MFE | -17.72 |
| Consensus MFE | -9.52 |
| Energy contribution | -9.58 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.50 |
| Mean z-score | -0.96 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.904572 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
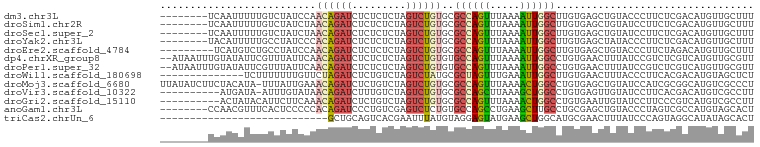
>dm3.chr3L 251981 91 - 24543557 --------UCAAUUUUUGUCUAUCCAACAGAUCUCUCUCUAGUCUGUGCGCCAGUUUAAAAUUGGCUUGUGAGCUGUACCCUUCUCGACAUGUUGCUUU --------.((((...((((......((((((.........))))))..((((((.....))))))....(((..(....)..))))))).)))).... ( -18.20, z-score = -1.31, R) >droSim1.chr2R 8475680 91 - 19596830 --------UCAAUUUUUGUCUAUCUAACAGAUCUCUCUCUAGUCUGUGCGCCAGUUUAAAAUUGGCUUGUGAGCUGUAUCCUUCUCGACAUGUUGCUUU --------.((((...((((......((((((.........))))))..((((((.....))))))....(((..(....)..))))))).)))).... ( -18.20, z-score = -1.42, R) >droSec1.super_2 279427 91 - 7591821 --------UCAAUUUUUGUCUAUCUAACAGAUCUCUCUCUAGUCUGUGCGCCAGUUUAAAAUUGGCUUGUGAGCUGUAUCCUUCUCGACAUGUUGCUUU --------.((((...((((......((((((.........))))))..((((((.....))))))....(((..(....)..))))))).)))).... ( -18.20, z-score = -1.42, R) >droYak2.chr3L 230594 91 - 24197627 --------UACAUUUUUGCCUAUCCCACAGAUCUCUCUCUAGUCUGUGCGCCAGUUUAAAAUUGGCUUGUGAGCUAUACCCUUCUCGACAUGUUGCUUU --------.........((......(((((((.........))))))).((((((.....)))))).((((((..........))).)))....))... ( -15.60, z-score = -0.58, R) >droEre2.scaffold_4784 240760 90 - 25762168 ---------UCAUGUCUGCCUAUCCAACAGAUCUCUCUCUAGUCUGUGCGCCAGUUUAAAAUUGGCUUGUGAGCUGUACCCUUCUAGACAUGUUGCUUU ---------.((((((((........((((((.........))))))..((((((.....))))))..................))))))))....... ( -20.70, z-score = -1.44, R) >dp4.chrXR_group8 7829731 97 + 9212921 --AUAAUUUGUAUAUUCGUUUAUUCAACAGAUCUCUCUCUAGUCUGUGUGCCAGUUUAAAAUUGGCCUGUGAACUUUAUCCGUCUCGUCAUGUUGCGUU --...............((((((...((((((.........))))))..((((((.....))))))..))))))......(((..(.....)..))).. ( -14.80, z-score = -0.93, R) >droPer1.super_32 92972 97 - 992631 --AUAAUUUGUAUAUUCGUUUAUUCAACAGAUCUCUCUCUAGUCUGUGUGCCAGUUUAAAAUUGGCCUGUGAACUUUAUCCGUCUCGUCAUGUUGCGUU --...............((((((...((((((.........))))))..((((((.....))))))..))))))......(((..(.....)..))).. ( -14.80, z-score = -0.93, R) >droWil1.scaffold_180698 39849 85 + 11422946 --------------UCUUUUUUUGUUCUAGAUCUCUGUCUAGUCUAUGCGCUAGUUUGAAAUUGGCUUGUGAACUUUACCCUUCACGACAUGUAGCUCU --------------............((((((....)))))).....((((((((.....))))))(((((((........)))))))......))... ( -16.00, z-score = -0.88, R) >droMoj3.scaffold_6680 2405746 98 - 24764193 UUAUAUCUUCUACAUA-UUUAUUGAAACAGAUCUCUGUCUAGUCUGUGCGCCAGUUUAAAACUGGCCUGUGAGCUGUAUCCAUCGCGGCAUGUCGCCCU ............((((-.........((((((.........))))))..((((((.....)))))).)))).(((((.......))))).......... ( -20.80, z-score = -0.66, R) >droVir3.scaffold_10322 368798 88 - 456481 ----------AUGAUA-AUUUGUAUAACAGAUCUUUGUCUAGUCUGUGCGCCAGCUUAAAGCUGGCCUGUGAGUUGUAUCCUUCACGACAUGUCGCCUU ----------...(((-(((..((..(((((.((......)))))))..((((((.....)))))).))..))))))........(((....))).... ( -23.30, z-score = -2.00, R) >droGri2.scaffold_15110 24407482 89 + 24565398 ----------ACUAUACAUUCUUCAAACAGAUCUCUGUCUAGUCUGUGCGCCAGUUUAAAACUGGCCUGUGAAUUGUAUCCUUCCCGUCAUGUCGCCUU ----------...(((((...((((.((((((.........))))))..((((((.....))))))...)))).))))).................... ( -17.80, z-score = -2.28, R) >anoGam1.chr3L 20293513 91 - 41284009 --------CCAACGUUUCACUCCCCCACAGAUCCCUGUCGAGUCUCUGUGCCAGCCUGAAGCUUGCCUGCGAGCUGUACCCUAGUCGCCAUGUAGCACU --------.....((((((......((((((..((....).)..))))))......))))))((((..((((.(((.....)))))))...)))).... ( -18.50, z-score = 0.15, R) >triCas2.chrUn_6 689170 71 + 1398711 ----------------------------GCUGCAGUCACGAAUUUAUGUAGGAGUAUGAAGCUGGCAUGCGAACUUUAUCCCAGUAGGCAUAUAGCACU ----------------------------.(((((............)))))..(((((..(((((.(((.......))).)))))...)))))...... ( -13.40, z-score = 1.22, R) >consensus ________UCAAUAUUUGUCUAUCCAACAGAUCUCUCUCUAGUCUGUGCGCCAGUUUAAAAUUGGCCUGUGAGCUGUAUCCUUCUCGACAUGUUGCUUU ..........................((((((.........))))))..((((((.....))))))................................. ( -9.52 = -9.58 + 0.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:52:19 2011