
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 199,235 – 199,335 |
| Length | 100 |
| Max. P | 0.710510 |

| Location | 199,235 – 199,335 |
|---|---|
| Length | 100 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 67.81 |
| Shannon entropy | 0.63795 |
| G+C content | 0.59206 |
| Mean single sequence MFE | -40.12 |
| Consensus MFE | -15.27 |
| Energy contribution | -14.79 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.65 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.710510 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 199235 100 - 24543557 UGCAACACGAACGCCACCUGCUUGCCUCCUGCUUGCACUCACGCGCAUAGUAAGCGCGCCCUU-AAACGAGCGUGCUAUGUGCGA-------GCGGGAGGGCGCCUGU---------- .((.........((.....))...((((((((((((((....((.....)).(((((((.(..-....).)))))))..))))))-------))))))))))......---------- ( -41.90, z-score = -1.62, R) >droSim1.chr3L 165321 100 - 22553184 UGCAACACGAACGCCACCUGCUUGCCUCCUGCUUGCACUCACGCGCAUAGUAAGCGCGCCCUU-AAACGAGCGUGCUGUGUGCGA-------GCGGGAGGGCGACUGU---------- .((.........((.....))...((((((((((((((.(..((.....)).(((((((.(..-....).)))))))).))))))-------))))))))))......---------- ( -42.70, z-score = -1.73, R) >droSec1.super_2 221847 100 - 7591821 UGCAACACGAACGCCACCUGCUUGCCUCCUGCUUGCACUCACGCGCAUAGUAAGCGCGCCCUU-AAACGAGCGUGCUGUGUGCGA-------GCGGGAGGGCGACUGU---------- .((.........((.....))...((((((((((((((.(..((.....)).(((((((.(..-....).)))))))).))))))-------))))))))))......---------- ( -42.70, z-score = -1.73, R) >droYak2.chr3L 177426 100 - 24197627 UGCAACACGCACGCCACCUGCUUGCCUCCUGCUCGCACUUACGCGCACAGUAAGCGCGCCCUU-AAACGAGCGUGCUGCGUGCGA-------GCAGGAGGGCGCCUGU---------- .(((...(((..((.....))...((((((((((((((.(((.......)))(((((((.(..-....).)))))))..))))))-------)))))))))))..)))---------- ( -47.70, z-score = -2.40, R) >droEre2.scaffold_4784 187899 100 - 25762168 UGCAACACGCACGCCACCUGCUUGCCUCUUACUCGCACUCACGCGCAUAGUAAGCGCGCCCUU-AAACGAGCGUGCUGCGUGCGA-------GCGGGAGGGCGCCUGU---------- (((.....)))((((.((((((((((...(((((((......)))...))))(((((((.(..-....).)))))))..).))))-------)))))..)))).....---------- ( -40.80, z-score = -0.56, R) >droAna3.scaffold_13337 22728000 101 + 23293914 UCCGACACGCACGCCACCUGCUUGCCUCUUGCUCGCACCUUCGGCCGCAAUGAGCGCGCCCUU-AGGCGAGCGUGCCAAGAGGGA-------GCGAGAGGGCGCACGGC--------- .......((..((((..((((((.((((((((.(((.(((..((((((.....))).)))...-)))...))).).)))))))))-------)).))..))))..))..--------- ( -45.10, z-score = -0.88, R) >dp4.chrXR_group8 7752255 101 + 9212921 UGCAACACGCGCGCCACCUGCUUGCCUCCCGCUCGCACUCAUAGACAGAGUAAGCGCGCCCUUAAAACGAGCGUGCUGUGUGGGA-------GCGAGAAGGCGUGAGU---------- ......((.((((((.....(((((.((((((.(..((((.......)))).(((((((.(.......).)))))))).))))))-------)))))..)))))).))---------- ( -43.00, z-score = -2.17, R) >droPer1.super_32 14922 101 - 992631 UGCAACACGCGCGCCACCUGCUUGCCUCCCGCUCGCACUCAUAGACAGAGUAAGCGCGCCCUUAAAACGAGCGUGCUGUGUGGGA-------GCGAGAAGGCGUGAGU---------- ......((.((((((.....(((((.((((((.(..((((.......)))).(((((((.(.......).)))))))).))))))-------)))))..)))))).))---------- ( -43.00, z-score = -2.17, R) >droWil1.scaffold_180698 4909209 100 - 11422946 UAAAACACUCAAGCCACCUGUUUUUCUCUCGCUCGCACUAAGGACAGCAAUAAGCGCGCCCUUUUGGCGAGCGAG-UGAGACAAA-------ACGAGUGAGCGGUUGU---------- ............(((((.((((((((((((((((((...((((.(.((.....))..).))))...)))))))).-.)))).)))-------))).))).))......---------- ( -37.20, z-score = -3.00, R) >droVir3.scaffold_13049 554409 107 - 25233164 UGCAGCACGCAC----GCCAACACUAUCUCGCUCGCACCAAUGCCGUAAAUGCGCGCGCCCUUAAAACGAGCGUGCGAGAUAGAUGGA--UUUUGGGAUGGCAUUUAAUGUUU----- (((.....))).----((((.((((((((..(((((((....(((((....))).))((.(.......).)))))))))..)))))).--...))...))))...........----- ( -31.30, z-score = 0.04, R) >droMoj3.scaffold_6680 24102402 115 + 24764193 UGCAGCACGCAC---GGUUAAUUCCAUCUCUCUCACACCAAUGCCGCGAAUGCGCGCGCCCUUAAAACGAGCGUGCGAGAUAGAUGGAUUUUUUGGGAUGGCAUUUAAUGUGUUGUUU .(((((((((.(---.(..((.(((((((.((((...........((....))((((((.(.......).)))))))))).))))))).))..).)....)).......))))))).. ( -36.61, z-score = -1.19, R) >droGri2.scaffold_15110 1808042 109 - 24565398 UGUAAAACGCAC----GCCACCUGCCUCUCGCUCGCACCUAUAUCGCCAAUGCGCGCGCCCUUAAAACGAGCGUGCGAGAUAGAUAGA----UUCGGAUGGCAUUUAAUGUUGUGUG- .......(((((----((((.(((.((.((.(((((((.......((....))((.((.........)).)))))))))...)).)).----..))).))))((.....)).)))))- ( -29.40, z-score = 0.00, R) >consensus UGCAACACGCACGCCACCUGCUUGCCUCCCGCUCGCACUCACGCCCACAGUAAGCGCGCCCUU_AAACGAGCGUGCUGUGUGGGA_______GCGGGAGGGCGUCUGU__________ ........................(((((((.........((.......)).(((((((.(.......).)))))))................))))))).................. (-15.27 = -14.79 + -0.48)
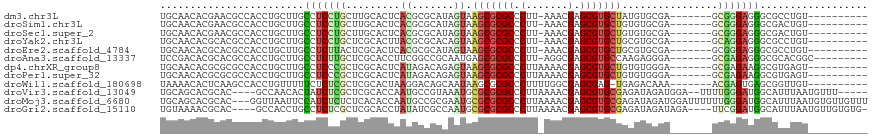

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:52:16 2011