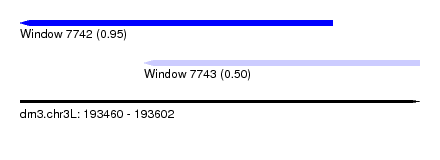
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 193,460 – 193,602 |
| Length | 142 |
| Max. P | 0.953618 |

| Location | 193,460 – 193,571 |
|---|---|
| Length | 111 |
| Sequences | 7 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 71.09 |
| Shannon entropy | 0.56686 |
| G+C content | 0.49914 |
| Mean single sequence MFE | -31.21 |
| Consensus MFE | -16.79 |
| Energy contribution | -16.90 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.64 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.953618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
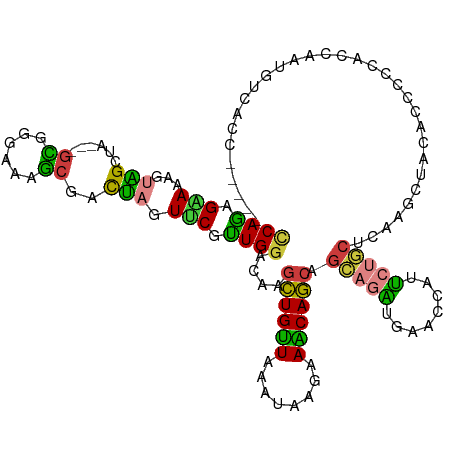
>dm3.chr3L 193460 111 - 24543557 CCAGAGAAAAGUAGCUA---GCGGAAAAGGGACUAGUUCGUUGGACAAGCUGUUAAAUAAGAAACAGCAGCAGAUGAACCAUUCUGCUCAAGCUACACCCCCACCAAUUUCACC----- ..........(((((((---((((((...((....(((.....)))..((((((........))))))..........)).)))))))..))))))..................----- ( -28.50, z-score = -2.24, R) >droVir3.scaffold_13049 546905 119 - 25233164 GCAGCGCAAGGCGGCGACGGGCGGGCGGGCGUCCAGUGCGCUGGAUAAGCUGCUCGACAAGAAGCAGCAGCAGCUCAACAAUGCCACGCCUGCGCCCACGGCAUCCACGGCAACCACGG .....((...(.((((.(((((((((((((((((((....))))))..(((((((.....).))))))....))))......))).)))))))))))...)).....((.......)). ( -51.80, z-score = -0.40, R) >droWil1.scaffold_180698 4901488 96 - 11422946 UCAAAGAAAGGCUGCAAGUGGAGGCAGAUCGAUAAGUGCAUUGGACAAAUUGUUGGAUCGCAAGCAACAGGAAUU---CAAUGCCCGGCCAGUGACACC-------------------- .........(((((...((.((......)).))....((((((((....((((((...(....)))))))...))---)))))).))))).........-------------------- ( -23.70, z-score = -0.18, R) >droEre2.scaffold_4784 182116 111 - 25762168 CCAGAGGAAAGUAGCCA---GUGGGAAAGCGACUAGUUCAUUGGACAAGCUGUUGAAUAAGAAACAGCAGCAGAUGAACCAUUCUGCACAGGCUACACCACCACCAAUGUCACC----- .....((...(((((((---((.(.....).))).(((.....)))..((((((........)))))).(((((........)))))...)))))).))...............----- ( -30.20, z-score = -1.83, R) >droYak2.chr3L 171643 111 - 24197627 CCAGAGGAAGGUAGCCA---GCGGAAAAGCGACUAGUUCGUUGGACAAGCUGUUGAAUAAGAAACAGCAGCAAUUGAACCAUGCUGCACAAGCUACACCACCACCAAUGUCACC----- .((..((..(((.((((---(((((..((...))..)))))))).).(((((((........))).((((((.........))))))...)))).....))).))..)).....----- ( -28.20, z-score = -1.16, R) >droSec1.super_2 216069 111 - 7591821 CCAGAGAAAAGUAGCUA---GCGGGAAAGCGACUAGUUCGUUGGACAAGCUGUUAAAUAAGAAACAGCAGCAGAUAAACCAUUCUGCUAAAGCUAAAUCCCCAUCAAUGUCACC----- ..........(.(((((---(((......)).)))))))....((((.((((((........))))))((((((........))))))...................))))...----- ( -27.10, z-score = -2.24, R) >droSim1.chr3L_random 4136 111 - 1049610 CCAGAGAAAAGUAGCUA---GCGGGAAAGCGACUAGUUCGUUGGACAAGCUGUUAAAUAAGAAACAGCAGCAGAUAAACCAUUCUGCUAAAGCUAAAUCCCCACCAAUGUCACC----- .....((.....(((((---(((......)).))))))((((((....((((((........))))))((((((........))))))...............))))))))...----- ( -29.00, z-score = -2.91, R) >consensus CCAGAGAAAAGUAGCUA___GCGGGAAAGCGACUAGUUCGUUGGACAAGCUGUUAAAUAAGAAACAGCAGCAGAUGAACCAUUCUGCUCAAGCUACACCCCCACCAAUGUCACC_____ ..........((((((....((((((......((((....))))....((((((........)))))).............))))))...))))))....................... (-16.79 = -16.90 + 0.11)

| Location | 193,504 – 193,602 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 72.45 |
| Shannon entropy | 0.49487 |
| G+C content | 0.45115 |
| Mean single sequence MFE | -25.03 |
| Consensus MFE | -12.03 |
| Energy contribution | -11.55 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.61 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
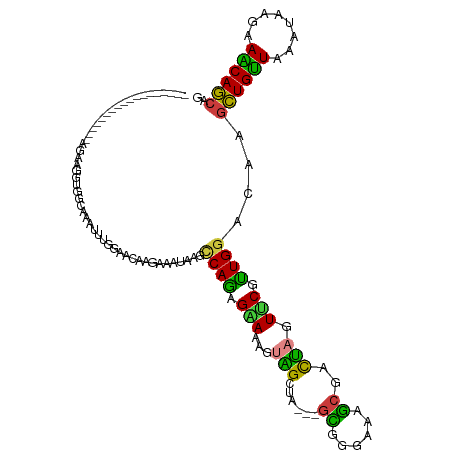
>dm3.chr3L 193504 98 - 24543557 ------------------AGAAGGUUGCAAAUUUGGAACAAGAAAUAAGCCAGAGAAAAGUAGCUA---GCGGAAAAGGGACUAGUUCGUUGGACAAGCUGUUAAAUAAGAAACAGCAG ------------------....((((((...(((((....(....)...))))).....))))))(---(((((..((...))..))))))......((((((........)))))).. ( -19.90, z-score = -1.68, R) >droVir3.scaffold_13049 546954 104 - 25233164 ---------------AGAAAACGCCGCCAAAUCUGGAGCAUGAGAUAGCGCAGCGCAAGGCGGCGACGGGCGGGCGGGCGUCCAGUGCGCUGGAUAAGCUGCUCGACAAGAAGCAGCAG ---------------......(((((((..((((........)))).(((...)))..))))))).(.((((.((((....)).)).)))).)....(((((((.....).)))))).. ( -42.00, z-score = -0.88, R) >droWil1.scaffold_180698 4901514 119 - 11422946 AAAAGGCGGCAUCAGCAGCAGUUCCAGCUAAUCUGGAAAUGGAGAUUAGUCAAAGAAAGGCUGCAAGUGGAGGCAGAUCGAUAAGUGCAUUGGACAAAUUGUUGGAUCGCAAGCAACAG .....(((((((..(((((..(((..((((((((........))))))))....)))..)))))..((.((......)).))..))))(((.(((.....))).))))))......... ( -31.80, z-score = -1.32, R) >droEre2.scaffold_4784 182160 98 - 25762168 ------------------AGAAGGUGGCAAAUUUGGAACAAGAAAUAAGCCAGAGGAAAGUAGCCA---GUGGGAAAGCGACUAGUUCAUUGGACAAGCUGUUGAAUAAGAAACAGCAG ------------------......((((..((((........))))..))))..........((((---(((((..((...))..)))))))).)..((((((........)))))).. ( -21.60, z-score = -1.87, R) >droYak2.chr3L 171687 98 - 24197627 ------------------AGAAGGUGGCAAAUUUAGAACAAGAAAUAAGCCAGAGGAAGGUAGCCA---GCGGAAAAGCGACUAGUUCGUUGGACAAGCUGUUGAAUAAGAAACAGCAG ------------------......((((..((((........))))..))))..........((((---(((((..((...))..)))))))).)..((((((........)))))).. ( -23.70, z-score = -2.67, R) >droSec1.super_2 216113 98 - 7591821 ------------------AGAAGGUUGCAAAUUUAGAACAAGAAAUUAGCCAGAGAAAAGUAGCUA---GCGGGAAAGCGACUAGUUCGUUGGACAAGCUGUUAAAUAAGAAACAGCAG ------------------.....(((.(((.....((((.((....((((............))))---((......))..)).)))).))))))..((((((........)))))).. ( -18.10, z-score = -0.62, R) >droSim1.chr3L_random 4180 98 - 1049610 ------------------AGAAGGUUGCAAAUUUAGAACAAGAAAUUAGCCAGAGAAAAGUAGCUA---GCGGGAAAGCGACUAGUUCGUUGGACAAGCUGUUAAAUAAGAAACAGCAG ------------------.....(((.(((.....((((.((....((((............))))---((......))..)).)))).))))))..((((((........)))))).. ( -18.10, z-score = -0.62, R) >consensus __________________AGAAGGUGGCAAAUUUGGAACAAGAAAUAAGCCAGAGAAAAGUAGCUA___GCGGGAAAGCGACUAGUUCGUUGGACAAGCUGUUAAAUAAGAAACAGCAG .................................................((((.(((.(((.((.............)).)))..))).))))....((((((........)))))).. (-12.03 = -11.55 + -0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:52:13 2011