| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,011,281 – 4,011,341 |
| Length | 60 |
| Max. P | 0.733394 |

| Location | 4,011,281 – 4,011,341 |
|---|---|
| Length | 60 |
| Sequences | 11 |
| Columns | 62 |
| Reading direction | reverse |
| Mean pairwise identity | 92.05 |
| Shannon entropy | 0.16407 |
| G+C content | 0.30290 |
| Mean single sequence MFE | -9.95 |
| Consensus MFE | -9.09 |
| Energy contribution | -9.09 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.733394 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 4011281 60 - 23011544 --UAGGACAAUUUCUAAAUGGAAUUUAAUUUCCGGAUUAGAAUAGACAUUUCAUGCAUGCUG --.........((((((.(((((......)))))..)))))).................... ( -9.10, z-score = -0.80, R) >droSim1.chr2L 3972333 60 - 22036055 --UAGGACAAUUUCUAAAUGGAAUUUAAUUUCCGGAUUAGAAUAGACAUUUCAUGCAUGCUG --.........((((((.(((((......)))))..)))))).................... ( -9.10, z-score = -0.80, R) >droSec1.super_5 2114780 60 - 5866729 --UAGGACAAUUUCUAAAUGGAAUUUAAUUUCCGGAUUAGAAUAGACAUUUCAUGCAUGCUG --.........((((((.(((((......)))))..)))))).................... ( -9.10, z-score = -0.80, R) >droYak2.chr2L 4029598 60 - 22324452 --UAGGACAAUUUCUAAAUGGAAUUUAAUUUCCGGAUUAGAAUAGACAUUUCAUGCAUGCUG --.........((((((.(((((......)))))..)))))).................... ( -9.10, z-score = -0.80, R) >droEre2.scaffold_4929 4070408 60 - 26641161 --UAGGACAAUUUCUAAAUGGAAUUUAAUUUCCGGAUUAGAAUAGACAUUUCAUGCAUGCUG --.........((((((.(((((......)))))..)))))).................... ( -9.10, z-score = -0.80, R) >droAna3.scaffold_12943 562426 62 + 5039921 UAUGGUACAAUUUCUAAAUGGAAUUUAAUUUCCGGAUUAGAAUAGACAUUUCGUGCAAGCUG ....((((...((((((.(((((......)))))..))))))..((....))))))...... ( -12.80, z-score = -2.06, R) >dp4.chr4_group3 9719246 60 + 11692001 --UAGGACAAUUUCUAAAUGGAAUUUAAUUUCCGGAUUAGAAUAGACAUUUCAUGCAUUUUG --.........((((((.(((((......)))))..)))))).................... ( -9.10, z-score = -1.19, R) >droPer1.super_1 6837015 60 + 10282868 --UAGGACAAUUUCUAAAUGGAAUUUAAUUUCCGGAUUAGAAUAGACAUUUCAUGCAUUUUG --.........((((((.(((((......)))))..)))))).................... ( -9.10, z-score = -1.19, R) >droWil1.scaffold_180703 1981401 60 - 3946847 --UGGGAAACUUUCUAAAUGGAAUUUAAUUUCCGGAUUAGAAUAGACAUUUCAUGCAAUUUG --.((....))((((((.(((((......)))))..)))))).................... ( -10.80, z-score = -1.47, R) >droVir3.scaffold_12963 10957551 59 + 20206255 ---UACGAAAUUUCUAAAUGGAAUUUAAUUUCCGGAUUAGAACAGACAUUUCAUGCAUUUUG ---...(((((((((((.(((((......)))))..)))))).....))))).......... ( -11.10, z-score = -2.59, R) >droMoj3.scaffold_6500 2064014 59 - 32352404 ---UACGAAAUUUCUAAAUGGAAUUUAAUUUCCGGAUUAGAAUAGACAUUUCAUGCAUUUUG ---...(((((((((((.(((((......)))))..)))))).....))))).......... ( -11.10, z-score = -2.34, R) >consensus __UAGGACAAUUUCUAAAUGGAAUUUAAUUUCCGGAUUAGAAUAGACAUUUCAUGCAUGCUG ...........((((((.(((((......)))))..)))))).................... ( -9.09 = -9.09 + 0.00)

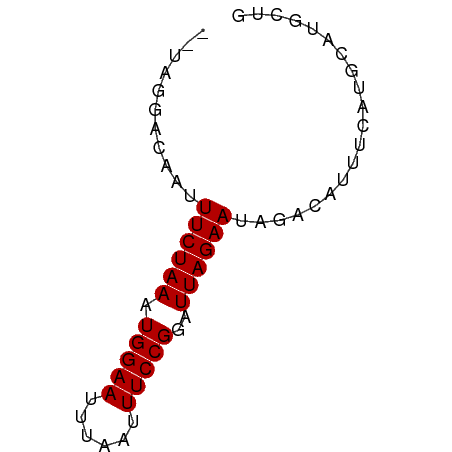

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:15:09 2011