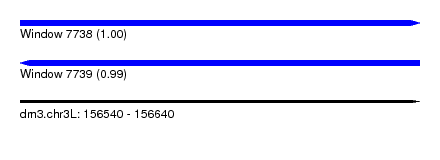
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 156,540 – 156,640 |
| Length | 100 |
| Max. P | 0.997475 |

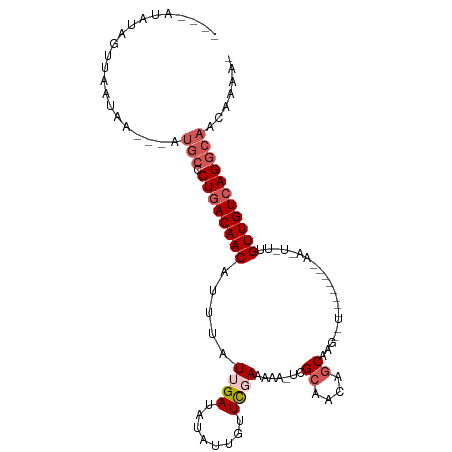
| Location | 156,540 – 156,640 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 73.68 |
| Shannon entropy | 0.42758 |
| G+C content | 0.32546 |
| Mean single sequence MFE | -20.94 |
| Consensus MFE | -12.48 |
| Energy contribution | -13.72 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.11 |
| SVM RNA-class probability | 0.997475 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
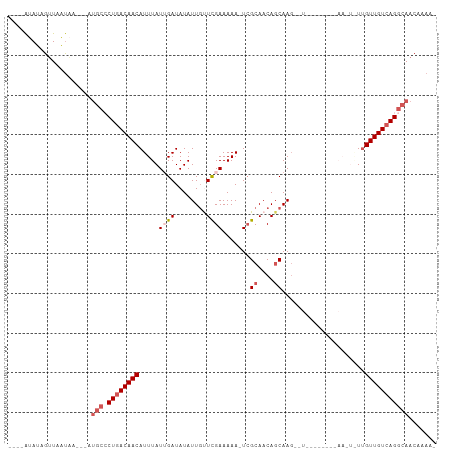
>dm3.chr3L 156540 100 + 24543557 ----AUAUAGUUAAUAA---AUGCCCUAACAACAUUUAUUGAUAUAUUCUUCGAAAAAAUCGCAACACCAACAAUCGCAACGCAACUAUCGUUGUCAGGCAACGAAU- ----.....((((((((---(((.........)))))))))))..((((..(((.....)))..............((...(((((....)))))...))...))))- ( -15.50, z-score = -1.81, R) >droEre2.scaffold_4784 137187 90 + 25762168 GUUCAAAAAGUUAUUCU--------CUGACAACAUUUAUGGAUAUAUUGCUUGAAAAA-UGGCAACAGCAAGUUU--------AAUUGUUGUUGUCAGGCAACAAAA- .(((((..(((...(((--------.(((......))).)))...)))..)))))...-((((((((((((....--------..))))))))))))(....)....- ( -22.10, z-score = -2.46, R) >droYak2.chr3L 128170 99 + 24197627 UUCCGAAAAGGUAUCCAUAAAUGCACUGACAACAUUUAUAGAUAUAUUGCUCGAAAAAAUCGCAACAGCAAGAUU--------AAUUGUUGUUGUCAGGCAACAAAA- ..((.....))..........(((.(((((((((...((.(((...((((((((.....)))....))))).)))--------.))...))))))))))))......- ( -24.10, z-score = -3.43, R) >droSec1.super_2 176140 84 + 7591821 ----AUAUAGUUAAUAA---AUGCCCUGACAACAUUUAUUGAUAUAUUGUUCGAAAAA-UCGCAACAGCAAG----------------UUGUUGUCAGGCAACUCAAU ----.............---.((((.((((((((....(((.....((((((((....-))).)))))))).----------------.))))))))))))....... ( -21.50, z-score = -3.65, R) >droSim1.chr3L 126195 84 + 22553184 ----AUAUAGUUAAUAA---AUGCCCUGACAACAUUUAUUGAUAUAUUGUUCGAAAAA-UCGCAACAGCAAG----------------UUGUUGUCAGGCAACUCAAU ----.............---.((((.((((((((....(((.....((((((((....-))).)))))))).----------------.))))))))))))....... ( -21.50, z-score = -3.65, R) >consensus ____AUAUAGUUAAUAA___AUGCCCUGACAACAUUUAUUGAUAUAUUGUUCGAAAAA_UCGCAACAGCAAG__U________AA_U_UUGUUGUCAGGCAACAAAA_ .....................(((.((((((((.....((((........)))).......((....)).....................)))))))))))....... (-12.48 = -13.72 + 1.24)

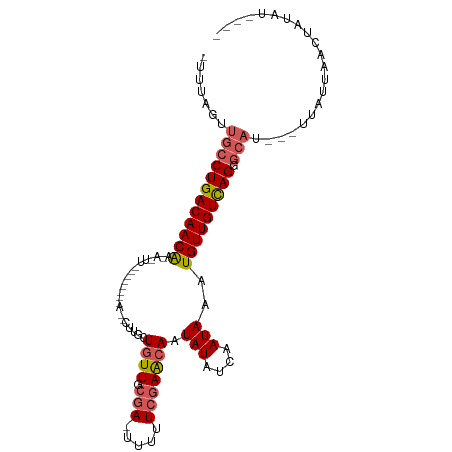
| Location | 156,540 – 156,640 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 73.68 |
| Shannon entropy | 0.42758 |
| G+C content | 0.32546 |
| Mean single sequence MFE | -21.82 |
| Consensus MFE | -13.68 |
| Energy contribution | -14.32 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.990139 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 156540 100 - 24543557 -AUUCGUUGCCUGACAACGAUAGUUGCGUUGCGAUUGUUGGUGUUGCGAUUUUUUCGAAGAAUAUAUCAAUAAAUGUUGUUAGGGCAU---UUAUUAACUAUAU---- -......((((((((((((((((((((...)))))))))(((((..(((.....)))......))))).......))))))).)))).---.............---- ( -24.90, z-score = -1.72, R) >droEre2.scaffold_4784 137187 90 - 25762168 -UUUUGUUGCCUGACAACAACAAUU--------AAACUUGCUGUUGCCA-UUUUUCAAGCAAUAUAUCCAUAAAUGUUGUCAG--------AGAAUAACUUUUUGAAC -..(((((..(((((((((.(((..--------....))).((((((..-........))))))..........)))))))))--------..))))).......... ( -17.60, z-score = -1.85, R) >droYak2.chr3L 128170 99 - 24197627 -UUUUGUUGCCUGACAACAACAAUU--------AAUCUUGCUGUUGCGAUUUUUUCGAGCAAUAUAUCUAUAAAUGUUGUCAGUGCAUUUAUGGAUACCUUUUCGGAA -...((((((....(((((.(((..--------....))).)))))(((.....))).))))))(((((((((((((.......)))))))))))))((.....)).. ( -25.80, z-score = -3.23, R) >droSec1.super_2 176140 84 - 7591821 AUUGAGUUGCCUGACAACAA----------------CUUGCUGUUGCGA-UUUUUCGAACAAUAUAUCAAUAAAUGUUGUCAGGGCAU---UUAUUAACUAUAU---- .((((..((((((((((((.----------------.....((((.(((-....))))))).(((....)))..)))))))).)))).---...))))......---- ( -20.40, z-score = -2.69, R) >droSim1.chr3L 126195 84 - 22553184 AUUGAGUUGCCUGACAACAA----------------CUUGCUGUUGCGA-UUUUUCGAACAAUAUAUCAAUAAAUGUUGUCAGGGCAU---UUAUUAACUAUAU---- .((((..((((((((((((.----------------.....((((.(((-....))))))).(((....)))..)))))))).)))).---...))))......---- ( -20.40, z-score = -2.69, R) >consensus _UUUAGUUGCCUGACAACAA_A_UU________A__CUUGCUGUUGCGA_UUUUUCGAACAAUAUAUCAAUAAAUGUUGUCAGGGCAU___UUAUUAACUAUAU____ .......((((((((((((......................((((.(((.....))))))).(((....)))..))))))))).)))..................... (-13.68 = -14.32 + 0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:52:10 2011