| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,005,113 – 4,005,215 |
| Length | 102 |
| Max. P | 0.905345 |

| Location | 4,005,113 – 4,005,215 |
|---|---|
| Length | 102 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.36 |
| Shannon entropy | 0.46459 |
| G+C content | 0.60305 |
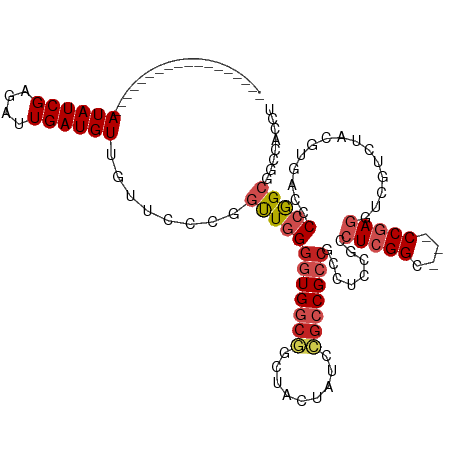
| Mean single sequence MFE | -42.27 |
| Consensus MFE | -22.36 |
| Energy contribution | -23.24 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.905345 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
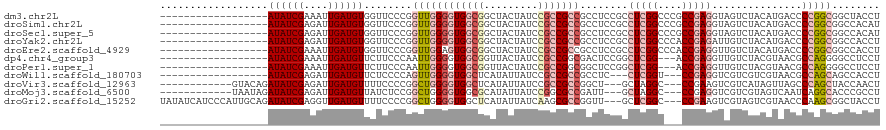
>dm3.chr2L 4005113 102 + 23011544 ------------------AUAUCGAAAUUGAUGUGGUUCCCGGUUGGGGUGGCGGCUACUAUCCGCCGCCGCCUCCGCCUCGGCCCGCCGAGGUAGUCUACAUGACCCCGGCGGCUACCU ------------------..............(((((..((((..(((((((((((........))))))))).))(((((((....))))))).(((.....))).))))..))))).. ( -52.00, z-score = -3.45, R) >droSim1.chr2L 3965236 102 + 22036055 ------------------AUAUCGAGAUUGAUGUGGUUCCCGGUUGGGGUGGCGGCUACUAUCCGCCGCCGCCUCCGCCUCGGCCCGCCGAGGUAGUCUACAUGACCCCGGCGGCCACAU ------------------............(((((((..((((..(((((((((((........))))))))).))(((((((....))))))).(((.....))).))))..))))))) ( -56.60, z-score = -4.27, R) >droSec1.super_5 2107608 102 + 5866729 ------------------AUAUCGAGAUUGAUGUGGUUCCCGGUUGGGGUGGCGGCUACUAUCCGCCGCCGCCUCCGCCUCGGCCCGCCGAGGUAGUCUACAUGACCCCGGCGGCCACAU ------------------............(((((((..((((..(((((((((((........))))))))).))(((((((....))))))).(((.....))).))))..))))))) ( -56.60, z-score = -4.27, R) >droYak2.chr2L 4022962 102 + 22324452 ------------------AUAUCGAGAUUGAUGUGGUUCCCGGUUGGGGUGGCGGCUACUAUCCGCCGCCGCCUCCGCCUCGGCCCACCGAGAUUGUCUACAUGACCCCGGCGGCCACCU ------------------..............(((((..((((..(((((((((((........))))))))).))..(((((....)))))...(((.....))).))))..))))).. ( -47.90, z-score = -2.83, R) >droEre2.scaffold_4929 4063867 102 + 26641161 ------------------AUAUCGAAAUUGAUGUGGUUCCCGGUUGGAGUGGCGGCUACUAUCCGCCGCCGCCUCCGCCUCGGCCCACCGAGGUUGUCUACAUGACCCCGGCGGCCACCU ------------------..............(((((..((((..(((((((((((........))))))).))))(((((((....))))))).(((.....))).))))..))))).. ( -50.10, z-score = -4.19, R) >dp4.chr4_group3 9711823 99 - 11692001 ------------------AUAUCGAAAUUGAUGUUCUUCCCAAUUGGGGUGGCGGUUACUAUCCGCCGGCGACUCCGGCUCGG---ACCGAGGUUGUCUACGUAACGCCAGGGGCCUCCU ------------------((((((....))))))....(((...(((.(((((((.......)))))((((((..(((.....---.)))..))))))......)).))).)))...... ( -34.70, z-score = -0.23, R) >droPer1.super_1 6829552 99 - 10282868 ------------------AUAUCGAAAUUGAUGUUCUUCCCAAUUGGGGUGGCGGUUACUAUCCGCCGGCGGCUCCGGCUCGG---ACCGAGGUUGUCUACGUAACGCCAGGGGCCUCCU ------------------((((((....))))))...((((....)))).(((((.......)))))((.((((((((((((.---..))).((((......))))))).)))))).)). ( -36.80, z-score = -0.43, R) >droWil1.scaffold_180703 1974453 96 + 3946847 ------------------AUAUCGAGAUUGAUGUUCUCCCCAGUUGGGGUGGCUCAUAUUAUCCGCCGCCGCCUC---CUCGGU---CCGAGGUCGUCGUCGUAACGCCAGCAGCCACCU ------------------.....((((.......))))....(((((((((((...........))))))((..(---((((..---.)))))..))..........)))))........ ( -27.90, z-score = 0.02, R) >droVir3.scaffold_12963 10946797 102 - 20206255 ------------GUACAGAUAUCGAGAUUGAUGUUUUCCCCGGCUGGGGUGGCUCAUAUUAUCCGCCGCCGGCU---GCUAGGC---CCGAAGUCGUCAUAGUUAGCCCAGCUACCAACU ------------....((((((((....)))))))).....(((((.((((((...........))))))((((---(((((((---........))).))).))))))))))....... ( -32.00, z-score = -0.54, R) >droMoj3.scaffold_6500 2047165 102 + 32352404 ------------UAAUAGAUAUCGAGAUUGAUGUUAUCUCCGGCUGGGGUGGCGCAUAUUAUCCGGCGCCGAUU---GCUAGGC---CCGAGGUCGUCGUAGUCAAUCAGGCACCCGCCU ------------...........(((((.......))))).(((.((((((((((..........)))))((((---((..(((---(...))))...))))))......)).)))))). ( -34.80, z-score = -0.32, R) >droGri2.scaffold_15252 12553626 114 + 17193109 UAUAUCAUCCCAUUGCAGAUAUCGAGGUUGAUGUUUUCCCCGGCUGGGGUGGCUCAUAUUAUCAAGCGCCGGUU---GCUCGGC---CCGAAGUCGUAGUCGUAACCCAAGCGGCUACCU .................(((.(((.((((((.((.....(((((...(((((......)))))....)))))..---)))))))---)))).)))((((((((.......)))))))).. ( -35.60, z-score = -0.34, R) >consensus __________________AUAUCGAGAUUGAUGUUGUUCCCGGUUGGGGUGGCGGCUACUAUCCGCCGCCGCCUCCGCCUCGGC___CCGAGGUCGUCUACGUGACCCCGGCGGCCACCU ..................((((((....))))))........((((((((((((.........)))))))........(((((....)))))...............)))))........ (-22.36 = -23.24 + 0.88)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:15:08 2011